Human longevity and Alzheimer's disease variants act via microglia and oligodendrocyte gene networks
- PMID: 39778705
- PMCID: PMC11884759
- DOI: 10.1093/brain/awae339
Human longevity and Alzheimer's disease variants act via microglia and oligodendrocyte gene networks
Abstract
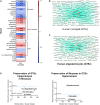
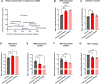
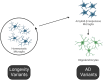
Ageing underlies functional decline of the brain and is the primary risk factor for several neurodegenerative conditions, including Alzheimer's disease (AD). However, the molecular mechanisms that cause functional decline of the brain during ageing, and how these contribute to AD pathogenesis, are not well understood. The objective of this study was to identify biological processes that are altered during ageing in the hippocampus and that modify Ad risk and lifespan, and then to identify putative gene drivers of these programmes. We integrated common human genetic variation associated with human lifespan or Ad from genome-wide association studies with co-expression transcriptome networks altered with age in the mouse and human hippocampus. Our work confirmed that genetic variation associated with Ad was enriched in gene networks expressed by microglia responding to ageing and revealed that they were also enriched in an oligodendrocytic gene network. Compellingly, longevity-associated genetic variation was enriched in a gene network expressed by homeostatic microglia whose expression declined with age. The genes driving this enrichment include CASP8 and STAT3, highlighting a potential role for these longevity-associated genes in the homeostatic functions of innate immune cells, and these genes might drive 'inflammageing'. Thus, we observed that gene variants contributing to ageing and AD balance different aspects of microglial and oligodendrocytic function. Furthermore, we also highlight putative Ad risk genes, such as LAPTM5, ITGAM and LILRB4, whose association with Ad falls below genome-wide significance but show strong co-expression with known Ad risk genes in these networks. Indeed, five of the putative risk genes highlighted by our analysis, ANKH, GRN, PLEKHA1, SNX1 and UNC5CL, have subsequently been identified as genome-wide significant risk genes in a subsequent genome-wide association study with larger sample size, validating our analysis. This work identifies new genes that influence ageing and AD pathogenesis, and highlights the importance of microglia and oligodendrocytes in the resilience of the brain against ageing and AD pathogenesis. Our findings have implications for developing markers indicating the physiological age of the brain and new targets for therapeutic intervention.
Keywords: Alzheimer’s disease; ageing; homeostasis; microglia; myelin; oligodendrocytes.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

