Investigating the origins of the mutational signatures in cancer
- PMID: 39778866
- PMCID: PMC11707540
- DOI: 10.1093/nar/gkae1303
Investigating the origins of the mutational signatures in cancer
Erratum in
-
Correction to 'Investigating the origins of the mutational signatures in cancer'.Nucleic Acids Res. 2025 Oct 28;53(20):gkaf1149. doi: 10.1093/nar/gkaf1149. Nucleic Acids Res. 2025. PMID: 41160902 Free PMC article. No abstract available.
Abstract
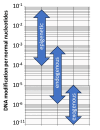
Most of the risk factors associated with chronic and complex diseases, such as cancer, stem from exogenous and endogenous exposures experienced throughout an individual's life, collectively known as the exposome. These exposures can modify DNA, which can subsequently lead to the somatic mutations found in all normal and tumor tissues. Understanding the precise origins of specific somatic mutations has been challenging due to multitude of DNA adducts (i.e. the DNA adductome) and their diverse positions within the genome. Thus far, this limitation has prevented researchers from precisely linking exposures to DNA adducts and DNA adducts to subsequent mutational outcomes. Indeed, many common mutations observed in human cancers appear to originate from error-prone endogenous processes. Consequently, it remains unclear whether these mutations result from exposure-induced DNA adducts, or arise indirectly from endogenous processes or are a combination of both. In this review, we summarize approaches that aim to bridge our understanding of the mechanism by which exposure leads to DNA damage and then to mutation and highlight some of the remaining challenges and shortcomings to fully supporting this paradigm. We emphasize the need to integrate cellular DNA adductomics, long read-based mapping, single-molecule duplex sequencing of native DNA molecules and advanced computational analysis. This proposed holistic approach aims to unveil the causal connections between key DNA modifications and the mutational landscape, whether they originate from external exposures, internal processes or a combination of both, thereby addressing key questions in cancer biology.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Abascal F., Harvey L.M.R., Mitchell E., Lawson A.R.J., Lensing S.V., Ellis P., Russell A.J.C., Alcantara R.E., Baez-Ortega A., Wang Y. et al. Somatic mutation landscapes at single-molecule resolution. Nature. 2021; 593:405–410. - PubMed
-
- Hollstein M., Sidransky D., Vogelstein B., Harris C.C. p53 mutations in human cancers. Science. 1991; 253:49–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM125503/GM/NIGMS NIH HHS/United States
- C66259/A27114/CRUK_/Cancer Research UK/United Kingdom
- S10 OD036306/OD/NIH HHS/United States
- P20 GM103429/GM/NIGMS NIH HHS/United States
- R01 ES032547/ES/NIEHS NIH HHS/United States
- R01 ES036931/ES/NIEHS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA269919/CA/NCI NIH HHS/United States
- Arkansas Research Alliance
- U01 CA290479/CA/NCI NIH HHS/United States
- NSTC 112-2314-B-040-013-MY3/National Science and Technology Council (Taiwan)
- R01 ES030557/ES/NIEHS NIH HHS/United States
- P20GM103429/NH/NIH HHS/United States
- OIA-1946391/National Science Foundation
- Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences
LinkOut - more resources
Full Text Sources
Medical

