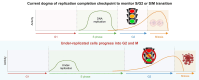
Cell cycle progression of under-replicated cells
- PMID: 39778868
- PMCID: PMC11707533
- DOI: 10.1093/nar/gkae1311
Cell cycle progression of under-replicated cells
Abstract
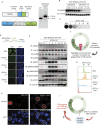
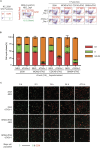
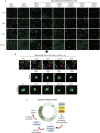
Cell cycle checkpoints are the regulatory mechanisms that secure the strict order of cellular events for cell division that ensure genome integrity. It has been proposed that mitosis initiation depends on the completion of DNA replication, which must be tightly controlled to guarantee genome duplication. Contrary to these conventional hypotheses, we showed here that cells were able to enter mitosis without completion of DNA replication. Although DNA replication was not completed in cells upon depletion of MCM2, CDC45 or GINS4, these under-replicated cells progressed into mitosis, which led to cell death. These unexpected results challenge current model and suggest the absence of a cell cycle checkpoint that monitors the completion of DNA replication.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Psychological therapies for panic disorder with or without agoraphobia in adults: a network meta-analysis.Cochrane Database Syst Rev. 2016 Apr 13;4(4):CD011004. doi: 10.1002/14651858.CD011004.pub2. Cochrane Database Syst Rev. 2016. PMID: 27071857 Free PMC article.
-
What is the value of routinely testing full blood count, electrolytes and urea, and pulmonary function tests before elective surgery in patients with no apparent clinical indication and in subgroups of patients with common comorbidities: a systematic review of the clinical and cost-effective literature.Health Technol Assess. 2012 Dec;16(50):i-xvi, 1-159. doi: 10.3310/hta16500. Health Technol Assess. 2012. PMID: 23302507 Free PMC article.
-
Histone Deacetylase Inhibitors Target DNA Replication Regulators and Replication Stress in Ewing Sarcoma Cells.Cancer Res Commun. 2025 Jun 1;5(6):1034-1048. doi: 10.1158/2767-9764.CRC-25-0058. Cancer Res Commun. 2025. PMID: 40478628 Free PMC article.
References
-
- Hartwell L.H., Weinert T.A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989; 246:629–634. - PubMed
-
- Elledge S.J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996; 274:1664–1672. - PubMed
-
- Torres-Rosell J., De Piccoli G., Cordon-Preciado V., Farmer S., Jarmuz A., Machin F., Pasero P., Lisby M., Haber J.E., Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007; 315:1411–1415. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

