Cortex-Specific Tmem169 Deficiency Induces Defects in Cortical Neuron Development and Autism-Like Behaviors in Mice
- PMID: 39779369
- PMCID: PMC11867004
- DOI: 10.1523/JNEUROSCI.1072-24.2024
Cortex-Specific Tmem169 Deficiency Induces Defects in Cortical Neuron Development and Autism-Like Behaviors in Mice
Abstract
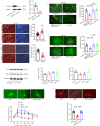
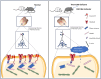
The development of the nervous system is a complex process, with many challenging scientific questions yet to be resolved. Disruptions in brain development are strongly associated with neurodevelopmental disorders, such as intellectual disability and autism. While the genetic basis of autism is well established, the precise pathological mechanisms remain unclear. Variations on chromosome 2q have been linked to autism, yet the specific genes responsible for the disorder have not been identified. This study investigates the role of the transmembrane protein 169 (TMEM169) gene, located on human chromosome 2q35, which has not been previously characterized. Our findings indicate that Tmem169 is highly expressed in the nervous system, and its deletion in the male mouse dorsal forebrain results in neuronal morphological abnormalities and synaptic dysfunction. Notably, Tmem169-deficient mice, irrespective of sex, display behavioral traits resembling those observed in individuals with autism. These results suggest that Tmem169 interacts with several key neuronal proteins, many of which are implicated in neurodevelopmental diseases. Furthermore, we demonstrate that Tmem169 promotes neuronal process and synapse development through its interaction with Shank3.
Keywords: Shank3; Tmem169; autism; brain development; neurodevelopmental disease.
Copyright © 2025 the authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Adhya D, et al. (2018) Atypical neurogenesis and excitatory-inhibitory progenitor generation in induced pluripotent stem cell (iPSC) from autistic individuals. bioRxiv, 349415.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
