Genetic code expansion reveals site-specific lactylation in living cells reshapes protein functions
- PMID: 39779673
- PMCID: PMC11711764
- DOI: 10.1038/s41467-024-55165-2
Genetic code expansion reveals site-specific lactylation in living cells reshapes protein functions
Abstract
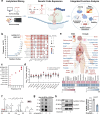
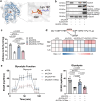
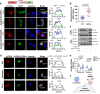
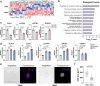
Protein lactylation is an emerging field. To advance the exploration of its biological functions, here we develop a comprehensive workflow that integrates proteomics to identify lactylated sites, genetic code expansion (GCE) for the expression of site-specifically lactylated proteins in living cells, and an integrated functional analysis (IFA) platform to evaluate their biological effects. Using a combined wet-and-dry-lab proteomics strategy, we identify a conserved lactylation at ALDOA-K147, which we hypothesize plays a significant biological role. Expression of this site-specifically lactylated ALDOA in mammalian cells reveals that this modification not only inhibits enzymatic activity but also induces gain-of-function effects. These effects reshaped ALDOA functionality by enhancing protein stability, promoting nuclear translocation, regulating adhesion-related gene expression, altering cell morphology and modulating ALDOA-interacting proteins. Our findings highlight the utility of the GCE-based workflow in establishing causal relationships between specific lactylation events and both target-specific and cell-wide changes, advancing our understanding of protein lactylation's functional impact.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 82173783/National Natural Science Foundation of China (National Science Foundation of China)
- 82322068/National Natural Science Foundation of China (National Science Foundation of China)
- 82404703/National Natural Science Foundation of China (National Science Foundation of China)
- 82104050/National Natural Science Foundation of China (National Science Foundation of China)
- 22377058/National Natural Science Foundation of China (National Science Foundation of China)
- BK20220088/Natural Science Foundation of Jiangsu Province (Jiangsu Provincial Natural Science Foundation)
- BK20241590/Natural Science Foundation of Jiangsu Province (Jiangsu Provincial Natural Science Foundation)
- BK20210692/Natural Science Foundation of Jiangsu Province (Jiangsu Provincial Natural Science Foundation)
- XPT82104050/Nanjing University of Chinese Medicine (NJUCM)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

