Pan-inhibition of super-enhancer-driven oncogenic transcription by next-generation synthetic ecteinascidins yields potent anti-cancer activity
- PMID: 39779693
- PMCID: PMC11711318
- DOI: 10.1038/s41467-024-55667-z
Pan-inhibition of super-enhancer-driven oncogenic transcription by next-generation synthetic ecteinascidins yields potent anti-cancer activity
Abstract
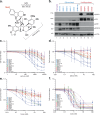
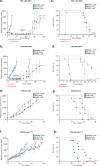
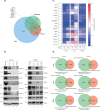
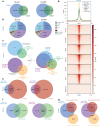
The plasticity of cancer cells facilitates their ability to adopt heterogeneous differentiation states, posing a significant challenge to therapeutic interventions. Specific gene expression programs, driven in part by super-enhancers (SEs), underlie cancer cell states. Here we successfully inhibit SE-driven transcription in phenotypically distinct metastatic melanoma cells using next-generation synthetic ecteinascidins. Through functional genomic methodologies, we demonstrate that these compounds inhibit the expression of genes encoding lineage-specific or ubiquitous transcription factors/coactivators by selectively targeting the CpG-rich sequences within their promoters and/or enhancers. This prevents the formation of transcription factor/coactivator condensates necessary for SE-dependent gene expression. Consequently, these compounds exhibit cytotoxic activity across distinct subpopulations of metastatic melanoma cells and inhibit tumor proliferation, including those resistant to current therapies. These findings extend to other cancers, like small cell lung cancer, recently approved for ecteinascidin-based treatment. Overall, our study provides preclinical proof that pan-inhibition of SE-dependent genes with synthetic ecteinascidins is a promising therapeutic approach for tumors with heterogeneous transcriptional landscapes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: PA and C-Cuevas are PharmaMar S.A employees and shareholders. MJGN, MMD, GSM are PharmaMar employees. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

