Identifying ADME-related gene signature for immune landscape and prognosis in KIRC by single-cell and spatial transcriptome analysis
- PMID: 39779746
- PMCID: PMC11711672
- DOI: 10.1038/s41598-024-84018-7
Identifying ADME-related gene signature for immune landscape and prognosis in KIRC by single-cell and spatial transcriptome analysis
Abstract
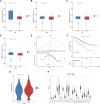
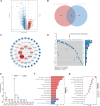
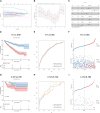
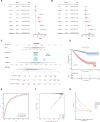
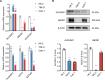
Kidney renal clear cell carcinoma (KIRC) is the most prevalent subtype of kidney cancer. Although multiple therapeutic agents have been proven effective in KIRC, their clinical application has been hindered by a lack of reliable biomarkers. This study focused on the prognostic value and function of drug absorption, distribution, metabolism, and excretion- (ADME-) related genes (ARGs) in KIRC to enhance personalized therapy. The critical role of ARGs in KIRC microenvironment was confirmed by single cell RNA-seq analysis and spatial transcriptome sequencing analysis for the first time. Then, an ADME-related prognostic signature (ARPS) was developed by the bulk RNA-seq analysis. The ARPS, created through Cox regression, LASSO, and stepAIC analyses, identified eight ARGs that stratified patients into high-risk and low-risk groups. High-risk patients had significantly poorer overall survival. Multivariate analysis confirmed the independent predictive ability of ARPS, and an ARPS-based nomogram was constructed for clinical application. Gene ontology and KEGG pathway analyses revealed immune-related functions and pathways enriched in these groups, with low-risk patients showing better responses to immunotherapy. Finally, the expression of ARGs was validated by qRT-PCR and Western blotting experiments. These findings underscore the prognostic significance of ARPS in KIRC and its potential application in guiding personalized treatment strategies.
Keywords: ADME genes; Gene signature; Immune cell infiltration; Kidney renal clear cell carcinoma; Survival.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Cell lines identity: The human kidney cell lines, including HK-2, 786-O, and Caki-1, were purchased from Wuhan Pricella Biotechnology Co.
Figures







References
-
- Li, Q. K., Pavlovich, C. P., Zhang, H., Kinsinger, C. R. & Chan, D. W. Challenges and opportunities in the proteomic characterization of clear cell renal cell carcinoma (ccRCC): A critical step towards the personalized care of renal cancers. Semin Cancer Biol55, 8–15. 10.1016/j.semcancer.2018.06.004 (2019). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- National Administration of Traditional Chinese Medicine talent education letter No. [2022]1/fifth batch of the National Traditional Chinese Medicine clinical outstanding talents training project
- 82074364/National Natural Science Foundation of China
- 82474412/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical

