Identification of a novel disulfidptosis-related gene signature in osteoarthritis using bioinformatics analysis and experimental validation
- PMID: 39779817
- PMCID: PMC11711464
- DOI: 10.1038/s41598-025-85569-z
Identification of a novel disulfidptosis-related gene signature in osteoarthritis using bioinformatics analysis and experimental validation
Abstract
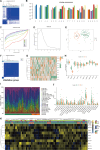
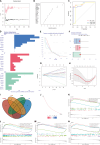
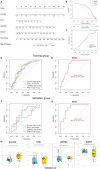
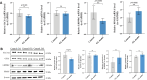
Osteoarthritis (OA) is a degenerative bone disease characterized by the destruction of joint cartilage and synovial inflammation, involving intricate immune regulation processes. Disulfidptosis, a novel form of programmed cell death, has recently been identified; however, the effects and roles of disulfidptosis-related genes (DR-DEGs) in OA remain unclear. We obtained six OA datasets from the GEO database, using four as training sets and two as validation sets. Differential expression analysis was employed to identify DR-DEGs, and unique molecular subtypes of OA were constructed based on these DR-DEGs. Subsequently, the immune microenvironment of OA patients was comprehensively analyzed using the "CIBERSORT" algorithm for immune infiltration. Various machine learning algorithms were utilized to screen characteristic DR-DEGs, and nomogram models and ROC curves were built based on these genes. The scRNA dataset (GSE169454) was used to classify chondrocytes in OA samples into distinct cell types, further exploring the gene distribution and correlation of characteristic DR-DEGs with specific cell subpopulations. Moreover, the expression levels of four characteristic DR-DEGs were validated through OA cell models and rat models. In our study, we identified 10 DR-DEGs with significant differences in expression within OA samples. Based on these DR-DEGs, two distinct molecular subtypes were recognized (cluster 1 and 2). ZNF484 and NDUFS1 were found to be significantly overexpressed in subtype 1, while the infiltration abundance of activated mast cells was markedly elevated in subtype 2. Moreover, significant differences were observed in the infiltration proportions of 11 immune cell types between OA and control samples, with 9 DR-DEGs demonstrating substantial correlations with immune cell infiltration levels. Further analysis of the scRNA dataset revealed that SLC3A2 and NDUFC1 were predominantly expressed in the preHTC subpopulation. All 10 DR-DEGs exhibited notably higher expression in the EC subpopulation across various cell types. The proportion of EC subgroups with high SLC3A2 expression increased, mainly enriching pathways related to inflammation, such as the IL-17 signaling pathway and TGF-beta signaling pathway. Using machine learning, we identified four characteristic DR-DEGs, which, in combination with the nomogram and ROC models, demonstrated promising performance in the diagnosis of OA. Additionally, in vivo validation confirmed a significant elevation of PPM1F expression in OA models. This study identified DR-DEGs as potential biomarkers for the diagnosis and classification of OA and provided a preliminary understanding of their role in the immune microenvironment. However, further experimental and clinical studies are required to validate their diagnostic value and therapeutic potential.
Keywords: Chondrocyte; Diagnosis; Disulfidptosis; Osteoarthritis; Single-cell sequencing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Informed consent: 920th Hospital of Joint Logistics Support Force Committee on Ethics approved this study and consented to participate (2024-053-01), and all methods are reported in accordance with ARRIVE guidelines.
Figures







Similar articles
-
Identification of osteoarthritis-associated chondrocyte subpopulations and key gene-regulating drugs based on multi-omics analysis.Sci Rep. 2025 Apr 11;15(1):12448. doi: 10.1038/s41598-025-90694-w. Sci Rep. 2025. PMID: 40216809 Free PMC article.
-
Developing the new diagnostic model by integrating bioinformatics and machine learning for osteoarthritis.J Orthop Surg Res. 2024 Dec 18;19(1):832. doi: 10.1186/s13018-024-05340-4. J Orthop Surg Res. 2024. PMID: 39695788 Free PMC article.
-
Systems biology-based analysis exploring shared biomarkers and pathogenesis of myocardial infarction combined with osteoarthritis.Front Immunol. 2024 Jul 17;15:1398990. doi: 10.3389/fimmu.2024.1398990. eCollection 2024. Front Immunol. 2024. PMID: 39086489 Free PMC article.
-
Identification and Construction of a Disulfidptosis-Mediated Diagnostic Model and Associated Immune Microenvironment of Osteoarthritis from the Perspective of PPPM.J Inflamm Res. 2024 Jun 11;17:3753-3770. doi: 10.2147/JIR.S462179. eCollection 2024. J Inflamm Res. 2024. PMID: 38882183 Free PMC article.
-
Chondrocyte channel transcriptomics: do microarray data fit with expression and functional data?Channels (Austin). 2013 Nov-Dec;7(6):459-67. doi: 10.4161/chan.26071. Epub 2013 Aug 30. Channels (Austin). 2013. PMID: 23995703 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

