Satellite DNA shapes dictate pericentromere packaging in female meiosis
- PMID: 39779853
- PMCID: PMC11880906
- DOI: 10.1038/s41586-024-08374-0
Satellite DNA shapes dictate pericentromere packaging in female meiosis
Abstract
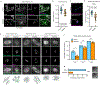
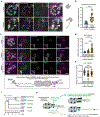
The abundance and sequence of satellite DNA at and around centromeres is evolving rapidly despite the highly conserved and essential process through which the centromere directs chromosome inheritance1-3. The impact of such rapid evolution is unclear. Here we find that sequence-dependent DNA shape dictates packaging of pericentromeric satellites in female meiosis through a conserved DNA-shape-recognizing chromatin architectural protein, high mobility group AT-hook 1 (HMGA1)4,5. Pericentromeric heterochromatin in two closely related mouse species, M. musculus and M. spretus, forms on divergent satellites that differ by both density of narrow DNA minor grooves and HMGA1 recruitment. HMGA1 binds preferentially to M. musculus satellites, and depletion in M. musculus oocytes causes massive stretching of pericentromeric satellites, disruption of kinetochore organization and delays in bipolar spindle assembly. In M. musculus × spretus hybrid oocytes, HMGA1 depletion disproportionately impairs M. musculus pericentromeres and microtubule attachment to their kinetochores. Thus, DNA shape affects both pericentromere packaging and the segregation machinery. We propose that rapid evolution of centromere and pericentromere DNA does not disrupt these essential processes when the satellites adopt DNA shapes recognized by conserved architectural proteins (such as HMGA1). By packaging these satellites, architectural proteins become part of the centromeric and pericentromeric chromatin, suggesting an evolutionary strategy that lowers the cost of megabase-scale satellite expansion.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








References
-
- Henikoff S, Ahmad K & Malik HS The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001). - PubMed
-
- Strauss F & Varshavsky A A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell 37, 889–901 (1984). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

