Functional evaluation and clinical classification of BRCA2 variants
- PMID: 39779857
- PMCID: PMC11821525
- DOI: 10.1038/s41586-024-08388-8
Functional evaluation and clinical classification of BRCA2 variants
Abstract
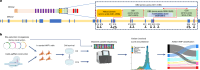
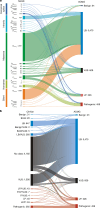
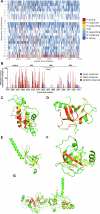
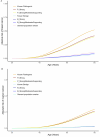
Germline BRCA2 loss-of function variants, which can be identified through clinical genetic testing, predispose to several cancers1-5. However, variants of uncertain significance limit the clinical utility of test results. Thus, there is a need for functional characterization and clinical classification of all BRCA2 variants to facilitate the clinical management of individuals with these variants. Here we analysed all possible single-nucleotide variants from exons 15 to 26 that encode the BRCA2 DNA-binding domain hotspot for pathogenic missense variants. To enable this, we used saturation genome editing CRISPR-Cas9-based knock-in endogenous targeting of human haploid HAP1 cells6. The assay was calibrated relative to nonsense and silent variants and was validated using pathogenic and benign standards from ClinVar and results from a homology-directed repair functional assay7. Variants (6,959 out of 6,960 evaluated) were assigned to seven categories of pathogenicity based on a VarCall Bayesian model8. Single-nucleotide variants that encode loss-of-function missense variants were associated with increased risks of breast cancer and ovarian cancer. The functional assay results were integrated into models from ClinGen, the American College of Medical Genetics and Genomics, and the Association for Molecular Pathology9 for clinical classification of BRCA2 variants. Using this approach, 91% were classified as pathogenic or likely pathogenic or as benign or likely benign. These classified variants can be used to improve clinical management of individuals with a BRCA2 variant.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.P., R.K. and M.R. are all employees of Ambry Genetics. All other authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

