A multi-kinase inhibitor screen identifies inhibitors preserving stem-cell-like chimeric antigen receptor T cells
- PMID: 39779871
- PMCID: PMC11785528
- DOI: 10.1038/s41590-024-02042-1
A multi-kinase inhibitor screen identifies inhibitors preserving stem-cell-like chimeric antigen receptor T cells
Abstract
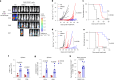
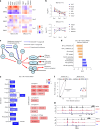
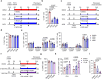
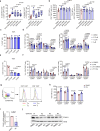
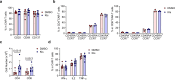
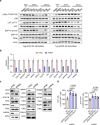
Chimeric antigen receptor T cells (CAR T cells) with T stem (TSCM) cell-like phenotypic characteristics promote sustained antitumor effects. We performed an unbiased and automated high-throughput screen of a kinase-focused compound set to identify kinase inhibitors (KIs) that preserve human TSCM cell-like CAR T cells. We identified three KIs, UNC10225387B, UNC10225263A and UNC10112761A, that combined in vitro increased the frequency of CD45RA+CCR7+TCF1hi TSCM cell-like CAR T cells from both healthy donors and patients with cancer. KI-treated CAR T cells showed enhanced antitumor effects both in vitro and in vivo in mouse tumor models. The KI cocktail maintains TSCM cell-like phenotype preferentially in CAR T cells originating from naive T cells and causes transcriptomic changes without arresting T cell activation or modulating the chromatin organization. Specific kinases, ITK, ADCK3, MAP3K4 and CDK13, targeted by the KI cocktail in a dose-dependent manner are directly associated with the preservation of TSCM cell-like CAR T cells. Knockdown of these kinases individually or in combination enriches for TSCM cell-like CAR T cells, but only CAR T cells generated in the presence of the KI cocktail show robust expansion and differentiation on stimulation with tumor cells. Overall, transient pharmacological inhibition of strategically targeted kinases maintains stem-like features in CAR T cells and improves their antitumor activity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: G.D. serves in the SAB of NanoCells, Estella, Arovella and Outspace Bio and is cofounder of Persistence Bio. A patent application has been submitted for the use of the identified KIs. Y.X. consults for Lumosa Therapeutics. The remaining authors declared no potential competing interests.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

