hsa_circ_0008305 facilitates the malignant progression of hepatocellular carcinoma by regulating AKR1C3 expression and sponging miR-379-5p
- PMID: 39779892
- PMCID: PMC11711494
- DOI: 10.1038/s41598-025-85737-1
hsa_circ_0008305 facilitates the malignant progression of hepatocellular carcinoma by regulating AKR1C3 expression and sponging miR-379-5p
Erratum in
-
Author Correction: hsa_circ_0008305 facilitates the malignant progression of hepatocellular carcinoma by regulating AKR1C3 expression and sponging miR-379-5p.Sci Rep. 2025 Sep 2;15(1):32311. doi: 10.1038/s41598-025-13503-4. Sci Rep. 2025. PMID: 40897744 Free PMC article. No abstract available.
Abstract
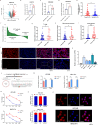
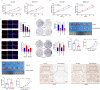
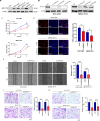
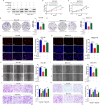
Circular RNAs (circRNAs) are widely involved in diverse biological processes of cancers. Nonetheless, the potential function of hsa_circ_0008305 in hepatocellular carcinoma (HCC) remains largely unknown. This study aims to elucidate the role and underlying mechanism of hsa_circ_0008305 in HCC. Our findings reveal that the novel circRNA hsa_circ_0008305 (circPTK2) is significantly upregulated in HCC tissues, with its elevated expression being positively correlated with advanced tumor T stage and vascular invasion. The circular characteristics and subcellular localization of hsa_circ_0008305 was determined by RNase R treatment and RNA nucleocytoplasmic separation. Further functional assays, including CCK8, EdU, colony formation assays, scratch-healing, transwell assays, and Xenograft tumor models were conducted to explore the biological functions of circPTK2. The regulatory mechanisms of circPTK2 were elucidated through RNA sequencing, enrichment analysis, and dual luciferase reporter assay. Our findings indicate that circPTK2 is stably localized in the cytoplasm. Functionally, circPKT2 promoted the HCC cells proliferation, migration, and invasion both in vitro and vivo. Mechanistically, circPTK2 was found to positively regulates the expression of AKR1C3 by acting as a sponge for miR-379-5p. Inhibition of miR-379-5p significantly mitigates the biological effects induced by circPTK2. AKR1C3 is identified as a direct target of miR-379-5p, and silencing AKR1C3 overturns the promotion progression effects of miR-379-5p inhibitor. In conclusion, our results revealed that circPTK2 facilitates the malignant progression of HCC via sponging miR-379-5p to up-regulate AKR1C3 expression.
Keywords: AKR1C3; Hepatocellular carcinoma; Hsa_circ_0008305; Tumor progression; miR-379-5p.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: All experiments with HCC tissues were approved by The Second Affiliated Hospital of Nanchang University Medical Research Ethics Committee, and the written informed consent was provided from each patient in accordance with the Helsinki Declaration. All animal experiments were approved by the Institutional Animal Care and Use Committee of Nanchang Royo Biotech Co,. Ltd. This study is conducted in accordance with ARRIVE guidelines. Consent for publication: Consent for publication has been obtained from the patients.
Figures




References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72 (1), 7–33 (2022). - PubMed
-
- Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71 (3), 209–249 (2021). - PubMed
-
- Parikh, N. D. & Pillai, A. Recent advances in hepatocellular carcinoma treatment. Clinical gastroenterology and hepatology: the official clinical practice. J. Am. Gastroenterol. Assoc.19 (10), 2020–2024 (2021). - PubMed
-
- Singal, A. G., Kanwal, F. & Llovet, J. M. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat. Rev. Clin. Oncol.20 (12), 864–884 (2023). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

