Chromosome mis-segregation triggers cell cycle arrest through a mechanosensitive nuclear envelope checkpoint
- PMID: 39779939
- PMCID: PMC11735390
- DOI: 10.1038/s41556-024-01565-x
Chromosome mis-segregation triggers cell cycle arrest through a mechanosensitive nuclear envelope checkpoint
Abstract
Errors during cell division lead to aneuploidy, which is associated with genomic instability and cell transformation. In response to aneuploidy, cells activate the tumour suppressor p53 to elicit a surveillance mechanism that halts proliferation and promotes senescence. The molecular sensors that trigger this checkpoint are unclear. Here, using a tunable system of chromosome mis-segregation, we show that mitotic errors trigger nuclear deformation, nuclear softening, and lamin and heterochromatin alterations, leading to rapid p53/p21 activation upon mitotic exit in response to changes in nuclear mechanics. We identify mTORC2 and ATR as nuclear deformation sensors upstream of p53/p21 activation. While triggering mitotic arrest, the chromosome mis-segregation-induced alterations of nuclear envelope mechanics provide a fitness advantage for aneuploid cells by promoting nuclear deformation resilience and enhancing pro-invasive capabilities. Collectively, this work identifies a nuclear mechanical checkpoint triggered by altered chromatin organization that probably plays a critical role in cellular transformation and cancer progression.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



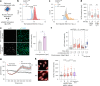

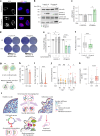


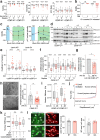



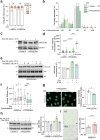
References
-
- Santaguida, S. & Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol.16, 473–485 (2015). - PubMed
-
- Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet.13, 189–203 (2012). - PubMed
-
- Hafner, A., Bulyk, M. L., Jambhekar, A. & Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol.20, 199–210 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

