H3K36 methylation regulates cell plasticity and regeneration in the intestinal epithelium
- PMID: 39779942
- PMCID: PMC12342706
- DOI: 10.1038/s41556-024-01580-y
H3K36 methylation regulates cell plasticity and regeneration in the intestinal epithelium
Abstract
Plasticity is needed during development and homeostasis to generate diverse cell types from stem and progenitor cells. Following differentiation, plasticity must be restricted in specialized cells to maintain tissue integrity and function. For this reason, specialized cell identity is stable under homeostatic conditions; however, cells in some tissues regain plasticity during injury-induced regeneration. While precise gene expression controls these processes, the regulatory mechanisms that restrict or promote cell plasticity are poorly understood. Here we use the mouse small intestine as a model system to study cell plasticity. We find that H3K36 methylation reinforces expression of cell-type-associated genes to maintain specialized cell identity in intestinal epithelial cells. Depleting H3K36 methylation disrupts lineage commitment and activates regenerative gene expression. Correspondingly, we observe rapid and reversible remodelling of H3K36 methylation following injury-induced regeneration. These data suggest a fundamental role for H3K36 methylation in reinforcing specialized lineages and regulating cell plasticity and regeneration.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: B.D.S. is a cofounder of EpiCypher. The other authors declare no competing interests.
Figures











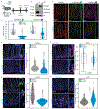





References
MeSH terms
Substances
Grants and funding
- R35GM142884/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- R35GM126900/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- P30 DK116073/DK/NIDDK NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- T32GM142607/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- F32DK135241-01/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- R35GM142884-02S/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- F32 DK135241/DK/NIDDK NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- R35 GM126900/GM/NIGMS NIH HHS/United States
- R35 GM142884/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

