Pancreatic organogenesis mapped through space and time
- PMID: 39779976
- PMCID: PMC11799519
- DOI: 10.1038/s12276-024-01384-y
Pancreatic organogenesis mapped through space and time
Abstract
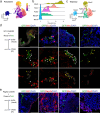
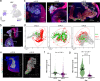
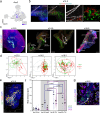
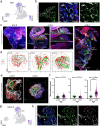
The spatial organization of cells within a tissue is dictated throughout dynamic developmental processes. We sought to understand whether cells geometrically coordinate with one another throughout development to achieve their organization. The pancreas is a complex cellular organ with a particular spatial organization. Signals from the mesenchyme, neurons, and endothelial cells instruct epithelial cell differentiation during pancreatic development. To understand the cellular diversity and spatial organization of the developing pancreatic niche, we mapped the spatial relationships between single cells over time. We found that four transcriptionally unique subtypes of mesenchyme in the developing pancreas spatially coordinate throughout development, with each subtype at fixed locations in space and time in relation to other cells, including beta cells, vasculature, and epithelial cells. Our work provides insight into the mechanisms of pancreatic development by showing that cells are organized in a space and time manner.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Generation of Scaffold-free, Three-dimensional Insulin Expressing Pancreatoids from Mouse Pancreatic Progenitors In Vitro.J Vis Exp. 2018 Jun 2;(136):57599. doi: 10.3791/57599. J Vis Exp. 2018. PMID: 29912186 Free PMC article.
-
Pancreatic mesenchyme regulates epithelial organogenesis throughout development.PLoS Biol. 2011 Sep;9(9):e1001143. doi: 10.1371/journal.pbio.1001143. Epub 2011 Sep 6. PLoS Biol. 2011. PMID: 21909240 Free PMC article.
-
Lineage dynamics of murine pancreatic development at single-cell resolution.Nat Commun. 2018 Sep 25;9(1):3922. doi: 10.1038/s41467-018-06176-3. Nat Commun. 2018. PMID: 30254276 Free PMC article.
-
Genes controlling pancreas ontogeny.Int J Dev Biol. 2008;52(7):823-35. doi: 10.1387/ijdb.072444cb. Int J Dev Biol. 2008. PMID: 18956314 Review.
-
Transcriptional regulation of pancreas development and β-cell function [Review].Endocr J. 2017 May 30;64(5):477-486. doi: 10.1507/endocrj.EJ17-0098. Epub 2017 Apr 14. Endocr J. 2017. PMID: 28420858 Review.
References
-
- Golosow, N. & Grobstein, C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Dev. Biol.4, 242–255 (1962). - PubMed
-
- Duvillié, B. et al. The mesenchyme controls the timing of pancreatic beta-cell differentiation. Diabetes55, 582–589 (2006). - PubMed
-
- Jacquemin, P. et al. An endothelial-mesenchymal relay pathway regulates early phases of pancreas development. Dev. Biol.290, 189–199 (2006). - PubMed
MeSH terms
Grants and funding
- UMO-2020/37/B/NZ3/01917/Narodowe Centrum Nauki (National Science Centre)
- UMO-2021/43/D/NZ3/02294/Narodowe Centrum Nauki (National Science Centre)
- POIR.04.04.00-00-20C5/16-00/Fundacja na rzecz Nauki Polskiej (Foundation for Polish Science)
- Druckenmiller Fellowship/New York Stem Cell Foundation (NYSCF)
- Hanna H. Gray Fellowship/Howard Hughes Medical Institute (HHMI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

