Exploring the role of splicing in TP53 variant pathogenicity through predictions and minigene assays
- PMID: 39780207
- PMCID: PMC11715486
- DOI: 10.1186/s40246-024-00714-5
Exploring the role of splicing in TP53 variant pathogenicity through predictions and minigene assays
Abstract
Background: TP53 variant classification benefits from the availability of large-scale functional data for missense variants generated using cDNA-based assays. However, absence of comprehensive splicing assay data for TP53 confounds the classification of the subset of predicted missense and synonymous variants that are also predicted to alter splicing. Our study aimed to generate and apply splicing assay data for a prioritised group of 59 TP53 predicted missense or synonymous variants that are also predicted to affect splicing by either SpliceAI or MaxEntScan.
Methods: We conducted splicing analyses using a minigene construct containing TP53 exons 2 to 9 transfected into human breast cancer SKBR3 cells, and compared results against different splice prediction methods, including correlation with the SpliceAI-10k calculator. We additionally applied the splicing results for TP53 variant classification using an approach consistent with the ClinGen Sequence Variant Interpretation Splicing Subgroup recommendations.
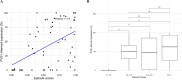
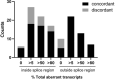
Results: Aberrant transcript profile consistent with loss of function, and for which a PVS1 (RNA) code would be assigned, was observed for 42 (71%) of prioritised variants, of which aberrant transcript expression was over 50% for 26 variants, and over 80% for 15 variants. Data supported the use of SpliceAI ≥ 0.2 cutoff for predicted splicing impact of TP53 variants. Prediction of aberration types using SpliceAI-10k calculator generally aligned with the corresponding assay results, though maximum SpliceAI score did not accurately predict level of aberrant expression. Application of the observed splicing results was used to reclassify 27/59 (46%) test variants as (likely) pathogenic or (likely) benign.
Conclusions: In conclusion, this study enhances the integration of splicing predictions and provides splicing assay data for exonic variants to support TP53 germline classification.
Keywords: PVS1; SpliceAI; Splicing; TP53; VCEP specifications.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This project has been approved by the QIMR Berghofer Medical Research Institute Human Research Ethics Committee (HREC) under QIMR HREC Approval P1051. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
-
- Fortuno C, Feng BJ, Carroll C, Innella G, Kohlmann W, Lázaro C, et al. Cancer risks associated with TP53 pathogenic variants: maximum likelihood analysis of extended pedigrees for diagnosis of first cancers beyond the Li-fraumeni syndrome spectrum. JCO Precis Oncol. 2024;8: e2300453. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- IIRS-21-102/National Breast Cancer Foundation
- APP177524/National Health and Medical Research Council
- APP177524/National Health and Medical Research Council
- ISCIII (PI20/00110)/Spanish Ministry of Science and Innovation, Acción Estratégica en Salud 2019
- ISCIII (PI24/00267)/Spanish Ministry of Science and Innovation, Acción Estratégica en Salud 2024
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

