Population sequencing of cherry accessions unravels the evolution of Cerasus species and the selection of genetic characteristics in edible cherries
- PMID: 39780235
- PMCID: PMC11708008
- DOI: 10.1186/s43897-024-00120-4
Population sequencing of cherry accessions unravels the evolution of Cerasus species and the selection of genetic characteristics in edible cherries
Erratum in
-
Correction: Population sequencing of cherry accessions unravels the evolution of Cerasus species and the selection of genetic characteristics in edible cherries.Mol Hortic. 2025 Feb 28;5(1):25. doi: 10.1186/s43897-025-00148-0. Mol Hortic. 2025. PMID: 40022255 Free PMC article. No abstract available.
Abstract
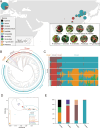
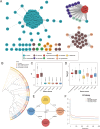
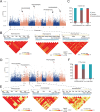
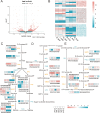
Cerasus is a subgenus of Prunus in the family Rosaceae that is popular owing to its ornamental, edible, and medicinal properties. Understanding the evolution of the Cerasus subgenus and identifying selective trait loci in edible cherries are crucial for the improvement of cherry cultivars to meet producer and consumer demands. In this study, we performed a de novo assembly of a chromosome-scale genome for the sweet cherry (Prunus avium L.) cultivar 'Burlat', covering 297.55 Mb and consisting of eight chromosomes with 33,756 protein-coding genes. The resequencing and population structural analysis of 384 Cerasus representative accessions revealed that they could be divided into four groups (Group 1, Group 2, Group 3, and Group 4). We inferred that Group 1 was the oldest population and Groups 2, 3, and 4 were clades derived from it. In addition, we found selective sweeps for fruit flavor and improved stress resistance in different varieties of edible cherries (P. avium, P. cerasus, and P. pseudocerasus). Transcriptome analysis revealed significant differential expression of genes associated with key pathways, such as sucrose starch and sucrose metabolism, fructose and mannose metabolism, and the pentose phosphate pathway, between the leaves and fruits of P. avium. This study enhances the understanding of the evolutionary processes of the Cerasus subgenus and provides resources for functional genomics research and the improvement of edible cherries.
Keywords: Cerasus subgenus; Edible cherries; Genetic diversity; Population structure; Selection signatures.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All authors approve the manuscript and consent to the publication of the work. Competing interests: The authors declare no competing interests. Caixi Zhang is a member of the Editorial Board for Molecular Horticulture. He was not involved in the journal’s review of, and decisions related to, this manuscript.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
