PRMT5-Mediated ALKBH5 Methylation Promotes Colorectal Cancer Immune Evasion via Increasing CD276 Expression
- PMID: 39781264
- PMCID: PMC11707101
- DOI: 10.34133/research.0549
PRMT5-Mediated ALKBH5 Methylation Promotes Colorectal Cancer Immune Evasion via Increasing CD276 Expression
Abstract
Numerous diseases have been connected to protein arginine methylations mediated by protein arginine methyltransferase 5 (PRMT5). Clinical investigations of the PRMT5-specific inhibitor GSK3326595 are currently being conducted, and the results are promising for preventing cancers. However, the detailed mechanism of PRMT5 promoting colorectal cancer (CRC) malignant progression remains unclear. Here, we found that PRMT5 directly catalyzes AlkB homologue 5 (ALKBH5) symmetric dimethylation at the R316 residue (meR316-ALKBH5), which enhances TRIM28-mediated ALKBH5 ubiquitination degradation. Then, an ALKBH5 decrease attenuates ALKBH5-mediated m6A demethylation on the CD276 transcript 3' untranslated region, which increases CD276 messenger RNA stability and its expression in CRC cells. Furthermore, a CD276 expression increase facilitates CRC immune evasion by inhibiting cytotoxic T-cell functions. Moreover, we revealed that PRMT5-mediated meR316-ALKBH5 activates CD276 transcription by increasing its messenger RNA m6A modification to increase CRC immune evasion in vitro and in vivo. Furthermore, we consistently showed a strong association between meR316-ALKBH5 and poor outcomes in patients with CRC. Finally, we demonstrated that combining an anti-PD1 antibody with the PRMT5 inhibitor GSK3326595 markedly halts the progression of CRC. Our findings could serve as a basis for the development of a PRMT5-meR316-ALKBH5-CD276 axis-targeting treatment approach for CRC.
Copyright © 2025 Sen Meng et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



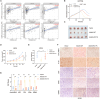


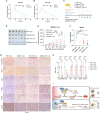
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. - PubMed
-
- Xu Y, Liu K, Li C, Li M, Liu F, Zhou X, Sun M, Ranganathan M, Zhang L, Wang S, et al. . The largest Chinese cohort study indicates homologous recombination pathway gene mutations as another major genetic risk factor for colorectal cancer with heterogeneous clinical phenotypes. Research (Wash D C). 2023;6: Article 0249. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

