The Cyclin-Dependent Kinase 8 Inhibitor E966-0530-45418 Attenuates Pulmonary Fibrosis In Vitro and In Vivo
- PMID: 39781457
- PMCID: PMC11705631
- DOI: 10.7150/ijbs.105826
The Cyclin-Dependent Kinase 8 Inhibitor E966-0530-45418 Attenuates Pulmonary Fibrosis In Vitro and In Vivo
Abstract
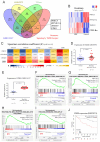
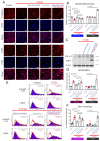
Pulmonary fibrosis (PF) is a high-mortality lung disease with limited treatment options, highlighting the need for new therapies. Cyclin-dependent kinase 8 (CDK8) is a promising target due to its role in regulating transcription via the TGF-β/Smad pathway, though CDK8 inhibitors have not been thoroughly studied for PF. This study aims to evaluate the potential of E966-0530-45418, a novel CDK8 inhibitor, in mitigating PF progression and explores its underlying mechanisms. We discovered that CDK8 is upregulated in lung tissues from idiopathic pulmonary fibrosis patients and in a bleomycin-induced PF mouse model. Our study further revealed that E966-0530-45418 inhibits PF progression by attenuating the activity of the transcription factor Smad3, which is involved in TGF-β1/Smad signaling, along with RNA polymerase II to downregulate fibrosis-associated protein expression in alveolar epithelia and lung fibroblasts and consequently mitigate myofibroblast differentiation and collagen deposition. E966-0530-45418 also blocks STAT3 signaling to obstruct M2 macrophage polarization, further suppressing PF progression. Moreover, E966-0530-45418 administration ameliorated lung function deterioration and lung parenchymal destruction in the bleomycin-induced PF mouse model. These findings indicate that E966-0530-45418 holds promise as a pioneering CDK8 inhibitor for treating PF.
Keywords: TGFβ1/Smad signaling; cyclin-dependent kinase 8; drug discovery; pulmonary fibrosis; transcriptional regulation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Lederer DJ, Martinez FJ. Idiopathic Pulmonary Fibrosis. N Engl J Med. 2018;378(19):1811–23. doi: https://doi.org/10.1056/NEJMra1705751. - PubMed
-
- Martinez FJ, Collard HR, Pardo A. et al. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers. 2017;3:17074. doi: https://doi.org/10.1038/nrdp.2017.74. - PubMed
-
- George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med. 2020;8(8):807–15. doi: https://doi.org/10.1016/S2213-2600(20)30225-3. - PMC - PubMed
-
- Kolahian S, Fernandez IE, Eickelberg O. et al. Immune Mechanisms in Pulmonary Fibrosis. Am J Respir Cell Mol Biol. 2016;55(3):309–22. doi: https://doi.org/10.1165/rcmb.2016-0121TR. - PubMed
-
- Edeling M, Ragi G, Huang S. et al. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol. 2016;12(7):426–39. doi: https://doi.org/10.1038/nrneph.2016.54. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

