Biological Amyloids Chemically Damage DNA
- PMID: 39782739
- PMCID: PMC11803820
- DOI: 10.1021/acschemneuro.4c00461
Biological Amyloids Chemically Damage DNA
Abstract
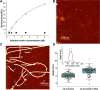
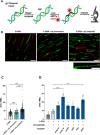
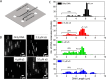
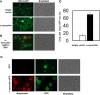
Amyloid fibrils are protein polymers noncovalently assembled through β-strands arranged in a cross-β structure. Biological amyloids were considered chemically inert until we and others recently demonstrated their ability to catalyze chemical reactions in vitro. To further explore the functional repertoire of amyloids, we here probe if fibrils of α-synuclein (αS) display chemical reactivity toward DNA. We demonstrate that αS amyloids bind DNA at micromolar concentrations in vitro. Using the activity of DNA repair enzymes as proxy for damage, we unravel that DNA-amyloid interactions promote chemical modifications, such as single-strand nicks, to the DNA. Double-strand breaks are also evident based on nanochannel analysis of individual long DNA molecules. The amyloid fold is essential for the activity as no DNA chemical modification is detected with αS monomers. In a yeast cell model, there is increased DNA damage when αS is overexpressed. Chemical perturbation of DNA adds another chemical reaction to the set of activities emerging for biological amyloids. Since αS amyloids are also found in the nuclei of neuronal cells of Parkinson's disease (PD) patients, and increased DNA damage is a hallmark of PD, we propose that αS amyloids contribute to PD by direct chemical perturbation of DNA.
Keywords: DNA damage; Parkinson’s disease; alpha-synuclein; amyloids; catalytic activity; nanochannels.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

