Large, recursive membrane platforms are associated to Trop-1, Trop-2, and protein kinase signaling for cell growth
- PMID: 39785844
- PMCID: PMC11974968
- DOI: 10.1091/mbc.E24-06-0267
Large, recursive membrane platforms are associated to Trop-1, Trop-2, and protein kinase signaling for cell growth
Abstract
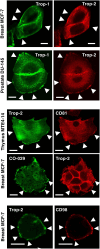
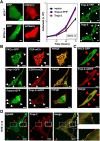
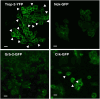
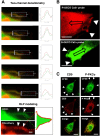
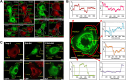
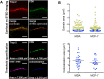
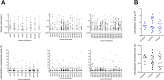
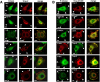
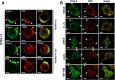
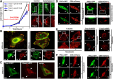
The transmembrane glycoproteins Trop-1/EpCAM and Trop-2 independently trigger Ca2+ and kinase signals for cell growth and tumor progression. Our findings indicated that Trop-1 and Trop-2 tightly colocalize at macroscopic, ruffle-like protrusions (RLP), that elevate from the cell perimeter, and locally recur over hundreds of seconds. These previously unrecognized elevated membrane regions ≥20-µm-long, up to 1.5 µm high were revealed by Z-stack analysis and three-dimensional reconstruction of signal transducer-hosting plasma membrane regions. Trop-2 stimulates cell growth through a membrane supercomplex that comprises CD9, PKCα, ion pumps, and cytoskeletal components. Our findings indicated that the growth-driving Trop-2 supercomplex assembles at RLP. RLP behaved as sites of clustering of signal transducers, of phosphorylation/activation of growth-driving kinases, as recruitment sites of PKCα and as origin of Ca2+ signaling waves, suggesting RLP to be novel signaling platforms in living cells. RLP were induced by growth factors and disappeared upon growth factor deprivation and β-actin depolymerization, candidating RLP to be functional platforms for high-dimensional signaling for cell growth.
Figures

References
-
- Alberti S (1998). A phosphoinositide-binding sequence is shared by PH domain target molecules–a model for the binding of PH domains to proteins. Proteins 31, 1–9. - PubMed
-
- Alberti S (1999). HIKE, a candidate protein binding site for PH domains, is a major regulatory region of Gbeta proteins. Proteins 35, 360–363. - PubMed
-
- Alberti S, Bucci C, Fornaro M, Robotti A, Stella M (1991). Immunofluorescence analysis in flow cytometry: Better selection of antibody-labeled cells after fluorescence overcompensation in the red channel. J Histochem Cytochem 39, 701–706. - PubMed
-
- Alberti S, Parks DR, Herzenberg LA (1987). A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry 8, 114–119. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

