Ultrasensitive Detection and Monitoring of Circulating Tumor DNA Using Structural Variants in Early-Stage Breast Cancer
- PMID: 39785866
- PMCID: PMC11994999
- DOI: 10.1158/1078-0432.CCR-24-3472
Ultrasensitive Detection and Monitoring of Circulating Tumor DNA Using Structural Variants in Early-Stage Breast Cancer
Abstract
Purpose: The detection of circulating tumor DNA (ctDNA) after curative-intent therapy in early-stage breast cancer is highly prognostic of disease recurrence. Current ctDNA assays, mainly targeting single-nucleotide variants, vary in sensitivity and specificity. Although increasing the number of single-nucleotide variants in tumor-informed assays improves sensitivity, structural variants (SV) may achieve similar or better sensitivity without compromising specificity. SVs occur across all cancers, linked to genomic instability and tumorigenesis, with unique tumor- and patient-specific breakpoints occurring throughout the genome. SVs in breast cancer are underexplored, and their potential for ctDNA detection and monitoring has not been fully evaluated.
Experimental design: We retrospectively analyzed a tumor-informed SV-based ctDNA assay in a cohort of patients with early-stage breast cancer (n = 100, 568 timepoints) receiving neoadjuvant systemic therapy, evaluating ctDNA dynamics and lead times to clinical recurrence in the postoperative period.
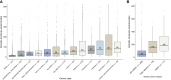
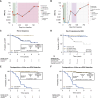
Results: ctDNA was detected in 96% (91/95) of participants at baseline with a median variant allele frequency of 0.15% (range: 0.0011%-38.7%); of these, 10% (9/91) had a variant allele frequency <0.01%. ctDNA detection at cycle 2 (C2) of neoadjuvant therapy was associated with a higher likelihood of distant recurrence (log-rank P = 0.047) and enhanced residual cancer burden prognostication (log-rank P = 0.041). ctDNA was detected prior to distant recurrence in all cases (100% sensitivity) with a median lead time of 417 days (range: 4-1,931 days).
Conclusions: These results demonstrate the clinical validity of ultrasensitive ctDNA detection and monitoring using SVs. Prospective trials are required to evaluate ctDNA-guided treatment strategies.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
K. Howarth reports other support from SAGA Dx during the conduct of the study as well as other support from NeoGenomics outside the submitted work. E. Amir reports personal fees from Pfizer and Novartis, as well as other support from Novartis outside the submitted work. M.B. Nadler reports personal fees from Novartis and Exact Sciences outside the submitted work. S. Bratman reports personal fees from Adela and EMD Serono and grants from AstraZeneca outside the submitted work as well as a patent for ctDNA mutation analysis issued, licensed, and with royalties paid from Roche and a patent for ctDNA methylation analysis issued, licensed, and with royalties paid from Adela. E.C. de Bruin reports being an AstraZeneca employee and holding AstraZeneca shares. C. Rushton reports personal fees from SAGA Dx during the conduct of the study. Y. Chen reports other support from SAGA Dx during the conduct of the study; in addition, Y. Chen has a patent for 63/650,048 pending to SAGA Dx, a patent for 63/650,061 pending to SAGA Dx, a patent for 63/497,872 pending to SAGA Dx, a patent for 63/402,511 pending to SAGA Dx, and a patent for 63/402,512 pending to SAGA Dx. S. Gladchuk reports other support from SAGA Dx during the conduct of the study as well as other support from SAGA Dx outside the submitted work; in addition, S. Gladchuk reports a patent for 63/402,512 pending to SAGA Dx. A.M. George reports other support from SAGA Dx during the conduct of the study; in addition, A.M. George has a patent for 63/497,872 pending, a patent for 18/240,416 pending, a patent for 63/650,048 pending, a patent for 63/650,052 pending, a patent for 63/650,061 pending, a patent for 63/402,511 pending, a patent for 63/402,512 pending, a patent for 63/348,855 pending, and a patent for 63/348,857 pending. S. Birkeälv reports other support from SAGA Dx during the conduct of the study as well as a patent for 18/240435 pending to SAGA Dx. M. Alcaide reports other support from SAGA Dx during the conduct of the study, as well as a patent for 63/497872 pending to SAGA Dx, 18/240416 pending to SAGA Dx, 63/650048 pending to SAGA Dx, 63/650052 pending to SAGA Dx, and 63/650061 pending to SAGA Dx. L. Oton reports other support from SAGA Dx during the conduct of the study and outside the submitted work, as well as a patent for 63/497872 pending to SAGA Dx, 63/650048 pending to SAGA Dx, and 63/650061 pending to SAGA Dx. G. Putcha reports personal fees from SAGA Dx during the conduct of the study and from Natera and Optum Genomics outside the submitted work. S. Woodhouse reports other support from Saga Dx during the conduct of the study, as well as patents 63/650048, 63/650052, and 63/650061 pending to SAGA Dx. P.L. Bedard reports grants from AstraZeneca, Bicara Therapeutics, Bayer, Boehringer Ingelheim, Merck, Novartis, Roche Genentech, LegoChem Biosciences, Medicenna, Zymeworks, Eli Lilly, Gilead, Takeda, GlaxoSmithKline, Bristol Myers Squibb, Amgen, and Pfizer outside the submitted work, as well as being Uncompensated Advisory for Janssen, Zymeworks, Repare Therapeutics, Lilly, Seagen, and Roche Genentech. L.L. Siu reports personal fees from Merck, Pfizer, AstraZeneca, Roche, GlaxoSmithKline, Voronoi, Arvinas, Navire, Relay, Daiichi Sankyo, Amgen, Marengo, Medicenna, Tubulis, LTZ Therapeutics, Pangea, and Break Through Cancer; grants from Novartis, Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, GlaxoSmithKline, Roche Genentech, AstraZeneca, Merck, Celgene, Astellas, Bayer, AbbVie, Amgen, Symphogen, Mirati, BioNTech, 23andMe, and EMD Serono; and personal fees and other support from Agios Pharmaceuticals and Treadwell Therapeutics outside the submitted work. D.W. Cescon reports financial support from AstraZeneca and other support from SAGA Dx during the conduct of the study as well as research support from Grail, Guardant Health, Inivata/NeoGenomics, Knight, and ProteinQure; personal fees and research support from AstraZeneca, GenomeRx, Gilead, GlaxoSmithKline, Merck, Pfizer, and Roche; and personal fees from Daiichi Sankyo, Lilly, Novartis, and SAGA Dx outside the submitted work. No disclosures were reported by the other authors.
Figures



References
-
- Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–48. - PubMed
-
- Donaldson J, Park BH. Circulating tumor DNA: measurement and clinical utility. Annu Rev Med 2018;69:223–34. - PubMed
-
- Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 2020;1:276–90. - PubMed
-
- Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res 2019;25:4255–63. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

