Human organoids for rapid validation of gene variants linked to cochlear malformations
- PMID: 39786576
- PMCID: PMC12003500
- DOI: 10.1007/s00439-024-02723-9
Human organoids for rapid validation of gene variants linked to cochlear malformations
Abstract
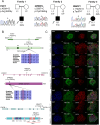
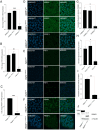
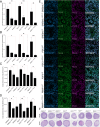
Developmental anomalies of the hearing organ, the cochlea, are diagnosed in approximately one-fourth of individuals with congenital. The majority of patients with cochlear malformations remain etiologically undiagnosed due to insufficient knowledge about underlying genes or the inability to make conclusive interpretations of identified genetic variants. We used exome sequencing for the genetic evaluation of hearing loss associated with cochlear malformations in three probands from unrelated families deafness. We subsequently generated monoclonal induced pluripotent stem cell (iPSC) lines, bearing patient-specific knockins and knockouts using CRISPR/Cas9 to assess pathogenicity of candidate variants. We detected FGF3 (p.Arg165Gly) and GREB1L (p.Cys186Arg), variants of uncertain significance in two recognized genes for deafness, and PBXIP1(p.Trp574*) in a candidate gene. Upon differentiation of iPSCs towards inner ear organoids, we observed developmental aberrations in knockout lines compared to their isogenic controls. Patient-specific single nucleotide variants (SNVs) showed similar abnormalities as the knockout lines, functionally supporting their causality in the observed phenotype. Therefore, we present human inner ear organoids as a potential tool to validate the pathogenicity of DNA variants associated with cochlear malformations.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

Update of
-
Human Organoids for Rapid Validation of Gene Variants Linked to Cochlear Malformations.Res Sq [Preprint]. 2024 Jun 11:rs.3.rs-4474071. doi: 10.21203/rs.3.rs-4474071/v1. Res Sq. 2024. Update in: Hum Genet. 2025 Apr;144(4):375-389. doi: 10.1007/s00439-024-02723-9. PMID: 38947059 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

