Unidirectional and bidirectional causation between smoking and blood DNA methylation: evidence from twin-based Mendelian randomisation
- PMID: 39786687
- PMCID: PMC11799127
- DOI: 10.1007/s10654-024-01187-5
Unidirectional and bidirectional causation between smoking and blood DNA methylation: evidence from twin-based Mendelian randomisation
Abstract
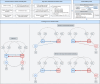
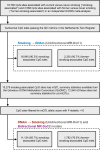
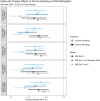
Cigarette smoking is associated with numerous differentially-methylated genomic loci in multiple human tissues. These associations are often assumed to reflect the causal effects of smoking on DNA methylation (DNAm), which may underpin some of the adverse health sequelae of smoking. However, prior causal analyses with Mendelian Randomisation (MR) have found limited support for such effects. Here, we apply an integrated approach combining MR with twin causal models to examine causality between smoking and blood DNAm in the Netherlands Twin Register (N = 2577). Analyses revealed potential causal effects of current smoking on DNAm at > 500 sites in/near genes enriched for functional pathways relevant to known biological effects of smoking (e.g., hemopoiesis, cell- and neuro-development, and immune regulation). Notably, we also found evidence of reverse and bidirectional causation at several DNAm sites, suggesting that variation in DNAm at these sites may influence smoking liability. Seventeen of the loci with putative effects of DNAm on smoking showed highly specific enrichment for gene-regulatory functional elements in the brain, while the top three sites annotated to genes involved in G protein-coupled receptor signalling and innate immune response. These novel findings are partly attributable to the analyses of current smoking in twin models, rather than lifetime smoking typically examined in MR studies, as well as the increased statistical power achieved using multiallelic/polygenic scores as instrumental variables while controlling for potential horizontal pleiotropy. This study highlights the value of twin studies with genotypic and DNAm data for investigating causal relationships of DNAm with health and disease.
Keywords: Causal inference; DNA Methylation; Epigenetics; Mendelian Randomisation; Smoking; Twin modelling.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflicts of interest: Nothing to declare.
Figures



Update of
-
Unidirectional and Bidirectional Causation between Smoking and Blood DNA Methylation: Evidence from Twin-based Mendelian Randomisation.medRxiv [Preprint]. 2024 Nov 22:2024.06.19.24309184. doi: 10.1101/2024.06.19.24309184. medRxiv. 2024. Update in: Eur J Epidemiol. 2025 Jan;40(1):55-69. doi: 10.1007/s10654-024-01187-5. PMID: 38946972 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

