NXP800 Activates the Unfolded Protein Response, Altering AR and E2F Function to Impact Castration-Resistant Prostate Cancer Growth
- PMID: 39787247
- PMCID: PMC11911806
- DOI: 10.1158/1078-0432.CCR-24-2386
NXP800 Activates the Unfolded Protein Response, Altering AR and E2F Function to Impact Castration-Resistant Prostate Cancer Growth
Abstract
Purpose: Advanced prostate cancer is invariably fatal, with the androgen receptor (AR) being a major therapeutic target. AR signaling inhibitors have improved overall survival for men with advanced prostate cancer, but treatment resistance is inevitable and includes reactivation of AR signaling. Novel therapeutic approaches targeting these mechanisms to block tumor growth is an urgent unmet clinical need. One attractive strategy is to target heat shock proteins (HSP) critical to AR functional activity.
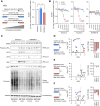
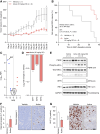
Experimental design: We first did transcriptome analysis on multiple castration-resistant prostate cancer (CRPC) cohorts to correlate the association between the Gene Ontology cellular response to heat gene expression signature and overall survival. Next, we analyzed the impact of targeting the heat shock factor 1 (HSF1) pathway, with an inhibitor in clinical development, namely, NXP800 (formerly CCT361814), in models of treatment-resistant prostate cancer. Finally, we confirmed our mechanistic and phenotypic findings using an NXP800-resistant model and an in vivo model of CRPC.
Results: We report that in multiple CRPC transcriptome cohorts, the Gene Ontology cellular response to heat gene expression signature associates with AR signaling and worse clinical outcome. We demonstrate the effects of targeting the HSF1 pathway, central to cellular stress, with an inhibitor in clinical development, namely, NXP800, in prostate cancer. Targeting the HSF1 pathway with the inhibitor NXP800 decreases HSP72 expression, activates the unfolded protein response, and inhibits AR- and E2F-mediated activity, inhibiting the growth of treatment-resistant prostate cancer models.
Conclusions: Overall, NXP800 has antitumor activity against treatment-resistant prostate cancer models, including molecular subtypes with limited treatment options, supporting its consideration for prostate cancer-specific clinical development.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
W. Zeng reports grants from the ICR during the conduct of the study. E. Pasqua reports personal fees from the ICR during the conduct of the study, as well as patents for WO201549535, WO2016156872, and WO2018065787 issued. N. Chessum reports personal fees from the ICR outside the submitted work, as well as a patent for WO2015049535 issued and licensed to Nuvectis Pharma. M. Cheeseman reports a patent for US11787786B2 issued to AstraZeneca UK Ltd, as well as patents for EP3283484A1 and EP3523295A1 issued to Cancer Research Technology Ltd. R. te Poele reports grants from Nuvectis Pharma and Sixth Element during the conduct of the study. M. Powers reports grants from Nuvectis Pharma and the ICR during the conduct of the study, as well as personal fees from the ICR outside the submitted work. P. Clarke reports grants from Nuvectis Pharma and Sixth Element during the conduct of the study. U. Banerji reports other support from Nuvectis Pharma during the conduct of the study; personal fees from Carrick Therapeutics, PharmEnable, Ellipsis Pharma, Dania Therapeutics, Amalus Therapeutics, and PEGASCY-GROUP; grants from Verastem Oncology and Avacta outside the submitted work; and a patent for the ICR issued. K. Jones reports grants from CRUK, Cancer Research Technology Pioneer Fund, and Battle Against Cancer Investment Trust during the conduct of the study, as well as a patent for WO2015/049535A1 issued, with royalties paid from Nuvectis Pharma. P. Workman reports personal fees from Alterome Therapeutics; grants and personal fees from Astex Pharmaceuticals, Merck KGaA, and Cyclacel Pharmaceuticals; other support from AstraZeneca, Chemical Probes Portal, CV6 Therapeutics, Epicombi Therapeutics, and Chroma Therapeutics; personal fees and other support from Nextech Invest, STORM Therapeutics, and Derwentwater Associates; grants, personal fees, and other support from Nuvectis Pharma; grants, nonfinancial support, and other support from Vivan Therapeutics; grants from Battle Against Cancer Investment Trust and Sixth Element Capital/CRT Pioneer Fund; grants and other support from the ICR; and personal fees and nonfinancial support from Nuevolution during the conduct of the study. P.S. Nelson reports grants from Janssen, as well as personal fees from Bristol Myers Squibb, Genentech, AstraZeneca, and Pfizer outside the submitted work. J.S. de Bono reports other support from AbbVie, Acai Therapeutics, Amgen, Astellas, Amunix, Bayer, Celcuity, Dark Blue Therapeutics, Duke Street Bio Ltd, GSK, Takeda, and Tango Therapeutics; personal fees from BioXcel Therapeutics, Daiichi, Dunad Therapeutics, Endeavor BioMedicines Inc., MacroGenics, MOMA Therapeutics, Nuvation Bio, One-carbon Therapeutics Inc., PAGE Therapeutics, and Tubulis GmbH; grants and other support from Crescendo, Genentech/Roche, Merck Serono, MetaCurUm, Myricx, Nurix Therapeutics, Oncternal Therapeutics, Orion Pharma, Sanofi, and Immunic Therapeutics; personal fees and other support from Novartis; and grants and personal fees from Pfizer, outside the submitted work; in addition, J.S. de Bono reports a patent for DNA damage repair inhibitors for treatment of cancer, licensed to AstraZeneca, and for 17-substituted steroids useful in cancer treatment, licensed to Janssen. A. Sharp reports other support from Sanofi, Roche Genentech, Nurix, Astellas Pharma, Merck Sharp & Dohme, DE Shaw Research, CHARM Therapeutics, Ellipses Pharma, and Droia Ventures outside the submitted work; being an employee of the ICR, which has a commercial interest in abiraterone, PARP inhibition in DNA repair defective cancers, and PI3K/AKT pathway inhibitors (no personal income); and being the CI/PI of industry-sponsored clinical trials. No disclosures were reported by the other authors.
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. - PubMed
-
- Westaby D, Fenor de La Maza MdLD, Paschalis A, Jimenez-Vacas JM, Welti J, de Bono J, et al. A new old target: androgen receptor signaling and advanced prostate cancer. Annu Rev Pharmacol Toxicol 2022;62:131–53. - PubMed
MeSH terms
Substances
Grants and funding
- Not known/Bone Cancer Research Trust (BCRT)
- R21 CA277368/CA/NCI NIH HHS/United States
- MR/W018217/1/Medical Research Council (MRC)
- C309/A11566/Cancer Research UK (CRUK)
- P50 CA097186/CA/NCI NIH HHS/United States
- R01 CA234715/CA/NCI NIH HHS/United States
- P50CA097186/National Institute of Health Sciences (NIHS)
- SGCL15/Academy of Medical Sciences (The Academy of Medical Sciences)
- RIA18-ST2-011/Prostate Cancer UK (ProstateUK)
- WT_/Wellcome Trust/United Kingdom
- Not known/CRIS Cancer Foundation (CRIS Foundation)
- CRUK CRM186x/Cancer Research UK (CRUK)
- R01 CA266452/CA/NCI NIH HHS/United States
- 219594/Z/19/Z/Wellcome Trust (WT)
- Not known/Chordoma Foundation (CF)
- N/A/NIHR Biomedical Research Centre, Royal Marsden NHS Foundation Trust/Institute of Cancer Research (BRC)
- 212969/Z/18/Z/Wellcome Trust (WT)
- R50 CA274336/CA/NCI NIH HHS/United States
- 21CHAL01/Prostate Cancer Foundation (PCF)
- TLD-PF19-006/Prostate Cancer UK (ProstateUK)
- Not known/Mark Foundation For Cancer Research (The Mark Foundation for Cancer Research)
- MR/M018318/1/Medical Research Council (MRC)
- P01 CA163227/CA/NCI NIH HHS/United States
- 18YOUN25/Prostate Cancer Foundation (PCF)
- R50CA274336/National Institute of Health Sciences (NIHS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

