Cu2+ mediates the oxidation of the transcription factor MscA to regulate the antioxidant defense of mycobacteria
- PMID: 39788544
- PMCID: PMC11711680
- DOI: 10.1093/nar/gkae1309
Cu2+ mediates the oxidation of the transcription factor MscA to regulate the antioxidant defense of mycobacteria
Abstract
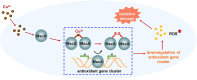
Copper (Cu), a trace element with redox activity, is both essential and toxic to living organisms. Its redox properties make it a cofactor for a variety of proteins, but it also causes oxidative stress, hence the need to maintain intracellular copper homeostasis. However, the role of copper in the regulation of antioxidant defense in bacteria remains unclear, and the involved transcription factors remain to be explored. In this study, we identified a novel transcription factor, MscA, that responded directly to Cu2+ to regulate the antioxidant defense of mycobacteria. Cu2+ directly bound to MscA to mediate oxidation and inhibit the DNA binding activity of MscA, subsequently downregulating the expression of antioxidant gene cluster to increase the accumulation of reactive oxygen species in mycobacteria, ultimately leading to oxidative damage to mycobacteria. Therefore, we firstly reported that the Cu2+ responsive transcription factor regulated the antioxidant defense in bacteria. This finding firstly and directly links the function of Cu2+ to the antioxidant defense of bacteria, and provides a new insight into bacterial antioxidant defense.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
Regulation of oxidative stress response and antioxidant modification in Corynebacterium glutamicum.World J Microbiol Biotechnol. 2024 Jul 15;40(9):267. doi: 10.1007/s11274-024-04066-z. World J Microbiol Biotechnol. 2024. PMID: 39004689 Review.
-
Expression of genes involved in redox homeostasis and antioxidant defense in a marine macroalga Ulva fasciata by excess copper.Aquat Toxicol. 2009 Oct 4;94(4):275-85. doi: 10.1016/j.aquatox.2009.07.010. Epub 2009 Jul 19. Aquat Toxicol. 2009. PMID: 19665240
-
Copper-caused oxidative stress triggers the activation of antioxidant enzymes via ZmMPK3 in maize leaves.PLoS One. 2018 Sep 17;13(9):e0203612. doi: 10.1371/journal.pone.0203612. eCollection 2018. PLoS One. 2018. PMID: 30222757 Free PMC article.
-
A conserved two-component system senses intracellular iron levels and regulates redox balance in Mycobacterium spp.Microbiol Spectr. 2024 Nov 5;12(11):e0110624. doi: 10.1128/spectrum.01106-24. Epub 2024 Sep 24. Microbiol Spectr. 2024. PMID: 39315783 Free PMC article.
-
Selected Trace Elements and Their Impact on Redox Homeostasis in Eye Health.Biomolecules. 2024 Oct 24;14(11):1356. doi: 10.3390/biom14111356. Biomolecules. 2024. PMID: 39595533 Free PMC article. Review.
References
-
- Furukawa Y., Shintani A., Kokubo T. A dual role of cysteine residues in the maturation of prokaryotic Cu/Zn-superoxide dismutase. Metallomics. 2021; 13:mfab050. - PubMed
-
- Frangipani E., Haas D. Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosaPAO1. FEMS MicrobioL Lett. 2009; 298:234–240. - PubMed
-
- Changela A., Chen K., Xue Y., Holschen J., Outten C.E., O’Halloran T.V., Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003; 301:1383–1387. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

