The role of ribosomal protein networks in ribosome dynamics
- PMID: 39788545
- PMCID: PMC11711686
- DOI: 10.1093/nar/gkae1308
The role of ribosomal protein networks in ribosome dynamics
Abstract
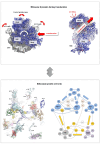
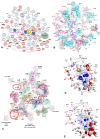
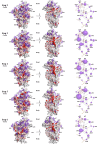
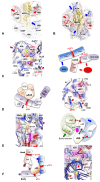
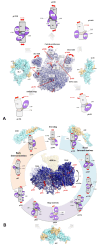
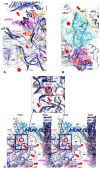
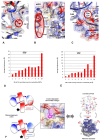
Accurate protein synthesis requires ribosomes to integrate signals from distant functional sites and execute complex dynamics. Despite advances in understanding ribosome structure and function, two key questions remain: how information is transmitted between these distant sites, and how ribosomal movements are synchronized? We recently highlighted the existence of ribosomal protein networks, likely evolved to participate in ribosome signaling. Here, we investigate the relationship between ribosomal protein networks and ribosome dynamics. Our findings show that major motion centers in the bacterial ribosome interact specifically with r-proteins, and that ribosomal RNA exhibits high mobility around each r-protein. This suggests that periodic electrostatic changes in the context of negatively charged residues (Glu and Asp) induce RNA-protein 'distance-approach' cycles, controlling key ribosomal movements during translocation. These charged residues play a critical role in modulating electrostatic repulsion between RNA and proteins, thus coordinating ribosomal dynamics. We propose that r-protein networks synchronize ribosomal dynamics through an 'electrostatic domino' effect, extending the concept of allostery to the regulation of movements within supramolecular assemblies.
Plain language summary
For accurate protein synthesis, ribosomes must coordinate signals and movements across their functional centers, but how this synchronization occurs remains unclear. Our study highlights the role of ribosomal protein networks in regulating ribosome dynamics. We show that major motion centers interact specifically with ribosomal proteins, and ribosomal RNA displays high mobility near negatively charged amino acids (Glu and Asp). Periodic electrostatic changes likely drive cycles of RNA–protein distance and approach, controlling key movements during translocation. Ribosomal protein networks may synchronize these dynamics through an ‘electrostatic domino effect,’ extending the concept of allostery to the regulation of molecular motions in supramolecular assemblies.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012; 19:560–567. - PubMed
-
- Bashan A., Yonath A. Correlating ribosome function with high-resolution structures. Trends Microbiol. 2008; 16:326–335. - PubMed
-
- Steitz T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008; 9:242–253. - PubMed
-
- Schmeing T.M., Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009; 461:1234–1242. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

