Induction of macrophage efferocytosis in pancreatic cancer via PI3Kγ inhibition and radiotherapy promotes tumour control
- PMID: 39788719
- PMCID: PMC12013568
- DOI: 10.1136/gutjnl-2024-333492
Induction of macrophage efferocytosis in pancreatic cancer via PI3Kγ inhibition and radiotherapy promotes tumour control
Abstract
Background: The immune suppression mechanisms in pancreatic ductal adenocarcinoma (PDAC) remain unknown, but preclinical studies have implicated macrophage-mediated immune tolerance. Hence, pathways that regulate macrophage phenotype are of strategic interest, with reprogramming strategies focusing on inhibitors of phosphoinositide 3-kinase-gamma (PI3Kγ) due to restricted immune cell expression. Inhibition of PI3Kγ alone is ineffective in PDAC, despite increased infiltration of CD8+ T cells.
Objective: We hypothesised that the immune stimulatory effects of radiation, and its ability to boost tumour antigen availability could synergise with PI3Kγ inhibition to augment antitumour immunity.
Design: We used orthoptic and genetically engineered mouse models of pancreatic cancer (LSL-KrasG12D/+;Trp53R172H/+;Pdx1-Cre). Stereotactic radiotherapy was delivered using contrast CT imaging, and PI3Kγ inhibitors by oral administration. Changes in the tumour microenvironment were quantified by flow cytometry, multiplex immunohistochemistry and RNA sequencing. Tumour-educated macrophages were used to investigate efferocytosis, antigen presentation and CD8+ T cell activation. Single-cell RNA sequencing data and fresh tumour samples with autologous macrophages to validate our findings.
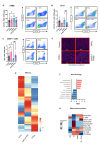
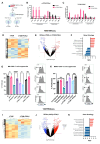
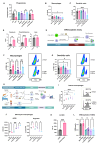
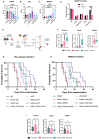
Results: Tumour-associated macrophages that employ efferocytosis to eradicate apoptotic cells can be redirected to present tumour antigens, stimulate CD8+ T cell responses and increase local tumour control. Specifically, we demonstrate how PI3Kγ signalling restricts inflammatory macrophages and that inhibition supports MERTK-dependent efferocytosis. We further find that the combination of PI3Kγ inhibition with targeted radiotherapy stimulates inflammatory macrophages to invoke a pathogen-induced like efferocytosis that switches from immune tolerant to antigen presenting.
Conclusions: Our data supports a new immunotherapeutic approach and a translational rationale to improve survival in PDAC.
Keywords: ANTIGEN PRESENTATION; CANCER; MACROPHAGES; PANCREATIC CANCER.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures



References
-
- Kaneda MM, Cappello P, Nguyen AV, et al. Macrophage PI3Kγ Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov. 2016;6:870–85. doi: 10.1158/2159-8290.CD-15-1346. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
