CDK4 inactivation inhibits apoptosis via mitochondria-ER contact remodeling in triple-negative breast cancer
- PMID: 39788939
- PMCID: PMC11718081
- DOI: 10.1038/s41467-024-55605-z
CDK4 inactivation inhibits apoptosis via mitochondria-ER contact remodeling in triple-negative breast cancer
Abstract
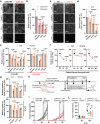
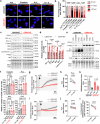
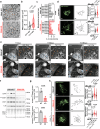
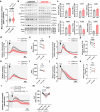
The energetic demands of proliferating cells during tumorigenesis require close coordination between the cell cycle and metabolism. While CDK4 is known for its role in cell proliferation, its metabolic function in cancer, particularly in triple-negative breast cancer (TNBC), remains unclear. Our study, using genetic and pharmacological approaches, reveals that CDK4 inactivation only modestly impacts TNBC cell proliferation and tumor formation. Notably, CDK4 depletion or long-term CDK4/6 inhibition confers resistance to apoptosis in TNBC cells. Mechanistically, CDK4 enhances mitochondria-endoplasmic reticulum contact (MERCs) formation, promoting mitochondrial fission and ER-mitochondrial calcium signaling, which are crucial for TNBC metabolic flexibility. Phosphoproteomic analysis identified CDK4's role in regulating PKA activity at MERCs. In this work, we highlight CDK4's role in mitochondrial apoptosis inhibition and suggest that targeting MERCs-associated metabolic shifts could enhance TNBC therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

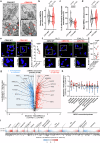

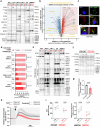
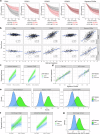

References
MeSH terms
Substances
Grants and funding
- 31003A_143369/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 310030_207688/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 8855/European Molecular Biology Organization (EMBO)
- J4597-B/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

