Enhancing immunotherapy efficacy in colorectal cancer: targeting the FGR-AKT-SP1-DKK1 axis with DCC-2036 (Rebastinib)
- PMID: 39788945
- PMCID: PMC11718245
- DOI: 10.1038/s41419-024-07263-8
Enhancing immunotherapy efficacy in colorectal cancer: targeting the FGR-AKT-SP1-DKK1 axis with DCC-2036 (Rebastinib)
Abstract
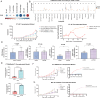
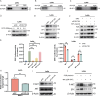
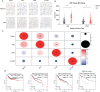
This research demonstrates that DCC-2036 (Rebastinib), a potent third-generation tyrosine kinase inhibitor (TKI), effectively suppresses tumor growth in colorectal cancer (CRC) models with functional immune systems. The findings underscore the capacity of DCC-2036 to enhance both the activation and cytotoxic functionality of CD8+ T cells, which are crucial for facilitating anti-tumor immune responses. Through comprehensive multi-omics investigations, significant shifts in both gene and protein expression profiles were detected, notably a marked decrease in DKK1 levels. This reduction in DKK1 was linked to diminished CD8+ T cell effectiveness, correlating with decreased FGR expression. Moreover, our findings identify FGR as a pivotal modulator that influences DKK1 expression via the PI3K-AKT-SP1 signaling cascade. Correlative analysis of clinical specimens supports the experimental data, showing that increased levels of FGR and DKK1 in CRC tissues are associated with inferior clinical outcomes and reduced efficacy of immunotherapeutic interventions. Consequently, targeting the FGR-AKT-SP1-DKK1 pathway with DCC-2036 could potentiate immunotherapy by enhancing CD8+ T cell functionality and their tumor infiltration. This strategy may contribute significantly to the refinement of therapeutic approaches for CRC, potentially improving patient prognoses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: No conflict of interest exists in the submission of this manuscript, and the manuscript is approved by all authors for publication. Ethics approval: The National Institutes of Health (NIH)‘s Guide for the Care and Use of Laboratory Animals served as the basis for this study. All efforts were made to minimize animal suffering during anesthesia with sodium pentobarbital (3.5% [w/v], 1 ml/kg). The experiments adhered to institutional guidelines and were approved by the ethics committee of the University of South China(# USC202209XS02). Consent for publication: Written informed consent for publication was obtained from all participants.
Figures





References
-
- Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233–54. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64. - PubMed
MeSH terms
Substances
Grants and funding
- 82103171/National Natural Science Foundation of China (National Science Foundation of China)
- 81972487/National Natural Science Foundation of China (National Science Foundation of China)
- 82473223/National Natural Science Foundation of China (National Science Foundation of China)
- 82271506/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

