Unveiling the molecular mechanisms of recurrent miscarriage through endoplasmic reticulum stress related gene expression
- PMID: 39789034
- PMCID: PMC11717954
- DOI: 10.1038/s41598-024-77642-w
Unveiling the molecular mechanisms of recurrent miscarriage through endoplasmic reticulum stress related gene expression
Abstract
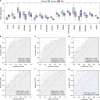
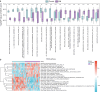
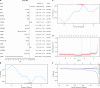
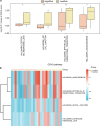
Recurrent miscarriage (RM) is a reproductive disorder affecting couples worldwide. The underlying molecular mechanisms remain elusive, even though emerging evidence has implicated endoplasmic reticulum stress (ERS). We investigated RM- and ERS-related genes to develop a diagnostic model that can enhance predictive ability. We utilized the R package GEO query to extract and process Gene Expression Omnibus data, applying batch correction, normalization, and differential gene expression analysis with limma. ERS-related differentially expressed genes (ERSRGs) were identified through Gene Ontology and Kyoto Encyclopedia of genes and genomes analyses, and their diagnostic potential was assessed. Diagnostic models were developed using logistic regression, support vector machines, and least absolute shrinkage and selection operators, complemented by immune infiltration analysis and regulatory network construction. Integrated analysis revealed 1395 differentially expressed genes (DEGs), including 626 upregulated and 769 downregulated genes. Seventeen ERSRGs were identified. KEAP1 and YIPF5 displayed high diagnostic accuracy (area under the curve [AUC] > 0.9). Gene Ontology and Kyoto Encyclopedia of genes and genomes analyses highlighted the role of ESRDEGs in cellular responses to ERS, protein processing, and apoptosis. Diagnostic models demonstrated robust predictive performance (AUC > 0.9). A molecular interaction was found between RM and the ERS response, and the identified ESRDEGs could serve as potential biomarkers for diagnosis.
Keywords: Bioinformatics; Diagnostic model; Endoplasmic reticulum stress; Immune Infiltration; Recurrent miscarriage.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
Endoplasmic reticulum stress and unfolded protein response play roles in recurrent pregnancy loss: A bioinformatics study.J Reprod Immunol. 2025 Mar;168:104446. doi: 10.1016/j.jri.2025.104446. Epub 2025 Feb 5. J Reprod Immunol. 2025. PMID: 39923360
-
Comprehensive analysis of endoplasmic reticulum stress-associated genes signature of ulcerative colitis.Front Immunol. 2023 May 23;14:1158648. doi: 10.3389/fimmu.2023.1158648. eCollection 2023. Front Immunol. 2023. PMID: 37287987 Free PMC article.
-
Identification and validation of endoplasmic reticulum stress-related genes in patients with steroid-induced osteonecrosis of the femoral head.Sci Rep. 2024 Sep 16;14(1):21634. doi: 10.1038/s41598-024-72941-8. Sci Rep. 2024. PMID: 39284931 Free PMC article.
-
Investigation and validation of genes associated with endoplasmic reticulum stress in diabetic retinopathy using various machine learning algorithms.Exp Eye Res. 2025 May;254:110317. doi: 10.1016/j.exer.2025.110317. Epub 2025 Mar 3. Exp Eye Res. 2025. PMID: 40043935
-
Integrative Analyses of Biomarkers Associated with Endoplasmic Reticulum Stress in Ischemic Stroke.Comput Math Methods Med. 2022 Aug 25;2022:4212180. doi: 10.1155/2022/4212180. eCollection 2022. Comput Math Methods Med. 2022. PMID: 36060663 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

