Serological insights from SARS-CoV-2 heterologous prime and boost responses in Thailand
- PMID: 39789037
- PMCID: PMC11718049
- DOI: 10.1038/s41598-024-84392-2
Serological insights from SARS-CoV-2 heterologous prime and boost responses in Thailand
Abstract
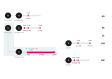
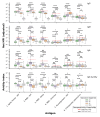
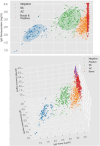
During the COVID-19 pandemic, heterologous vaccination strategies were employed to alleviate the strain on vaccine supplies. The Thailand Ministry of Health adopted these strategies using vector, inactivated, and mRNA vaccines. However, this approach has introduced challenges for SARS-CoV-2 sero-epidemiology studies. Our study analysed 647 samples from healthcare workers who received CoronaVac, ChAdOx1 nCoV-19, and BNT162b2 vaccines. The serological profile encompassed responses to various SARS-CoV-2 variants and vectors, measuring IgG, IgM, and IgA isotypes, alongside IgG avidity assays. The results demonstrated that heterologous CoronaVac/ChAdOx1 nCoV-19 schedules elicited significantly stronger antibody responses compared to homologous schedules (IgG: 1.2-fold, IgM: 10.9-fold, IgA: 3.1-fold increase). Additionally, a heterologous BNT162b2 boost at 4-weeks post-initial vaccination showed greater antibody levels than a ChAdOx1 nCoV-19 boost (IgG: 1.1-fold, IgM: slight decrease, IgA: 1.5-fold increase). Using a combination of three analytes, IgG against wild-type Spike trimer, nucleoprotein and Omicron receptor binding domains, enabled the clustering of responses within a statistical Gaussian mixture model that successfully discriminates between breakthrough infections and vaccination types (F-score = 0.82). The development of statistical models to predict breakthrough infections can improve serological surveillance. Overall, our study underscores the necessity for vaccine (re-)development and the creation of serological tools to monitor vaccine performance.
Keywords: Antibody; COVID-19; SARS-CoV-2; Vaccine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This study was approved by the research ethics committee, Department of Medical Sciences in October 2021 (ref. EC15/2564). Competing interests: The authors declare no competing interests.
Figures

References
-
- Sahin, U., Muik, A., Vogler, I., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021 595:7868 2021; 595: 572–7. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

