Ongoing replication stress tolerance and clonal T cell responses distinguish liver and lung recurrence and outcomes in pancreatic cancer
- PMID: 39789181
- PMCID: PMC11779630
- DOI: 10.1038/s43018-024-00881-3
Ongoing replication stress tolerance and clonal T cell responses distinguish liver and lung recurrence and outcomes in pancreatic cancer
Abstract
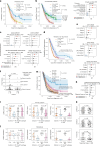
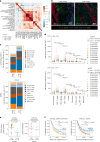
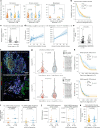
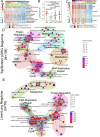
Patients with metastatic pancreatic ductal adenocarcinoma survive longer if disease spreads to the lung but not the liver. Here we generated overlapping, multi-omic datasets to identify molecular and cellular features that distinguish patients whose disease develops liver metastasis (liver cohort) from those whose disease develops lung metastasis without liver metastases (lung cohort). Lung cohort patients survived longer than liver cohort patients, despite sharing the same tumor subtype. We developed a primary organotropism (pORG) gene set enriched in liver cohort versus lung cohort primary tumors. We identified ongoing replication stress response pathways in high pORG/liver cohort tumors, whereas low pORG/lung cohort tumors had greater densities of lymphocytes and shared T cell clonal responses. Our study demonstrates that liver-avid pancreatic ductal adenocarcinoma is associated with tolerance to ongoing replication stress, limited tumor immunity and less-favorable outcomes, whereas low replication stress, lung-avid/liver-averse tumors are associated with active tumor immunity that may account for favorable outcomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Disclosures relevant to PDAC are as follows. R.C.S. declares Scientific Advisory Board (SAB)/consultancy for Rappta Therapeutics, PanCAN, PRECEDE, MOHCCN & PanCuRx Canada and Precision Panc CRUK; sponsored research for Cardiff Oncology, AstraZeneca. G.B.M. declares SAB/consultancy for Amphista, Astex, AstraZeneca, BlueDot, Chrysallis Biotechnology, Ellipses Pharma, ImmunoMET, Infinity, Ionis, Lilly, Medacorp, Nanostring, Nuvectis, PDX Pharmaceuticals, Roche, Signalchem Lifesciences, Tarveda, Turbine and Zentalis Pharmaceuticals; Stock/options/financial for AstraZeneca, Catena Pharmaceuticals, ImmunoMet, SignalChem, Tarveda and Turbine; licensed technology HRD assay to Myriad Genetics and Digital Spatial Profiler patents with Nanostring. L.M.C. declares consulting services for Cell Signaling Technologies, AbbVie, the Susan G Komen Foundation and Shasqi, received reagents and/or research support from Cell Signaling Technologies, Syndax Pharmaceuticals, ZelBio, Hibercell and Acerta Pharma, and participates in advisory boards for Pharmacyclics, Syndax, Carisma, Verseau, CytomX, Kineta, Hibercell, Cell Signaling Technologies, Alkermes, Zymeworks, Genenta Sciences, Pio Therapeutics, PDX Pharmaceuticals, NextCure, the AstraZeneca Partner of Choice Network, the Lustgarten Foundation and the NIH–NCI-Frederick National Laboratory Advisory Committee. J.R.B. declares SAB for Perthera, Advisory for IDEAYA and is an editor for Springer and Taylor & Francis publishing. The other authors declare no competing interests.
Figures











References
-
- He, C., Huang, X., Zhang, Y., Lin, X. & Li, S. The impact of different metastatic patterns on survival in patients with pancreatic cancer. Pancreatology21, 556–563 (2021). - PubMed
MeSH terms
Grants and funding
- U01CA278923/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01CA186241/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U01 CA253472/CA/NCI NIH HHS/United States
- U01CA253472/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- 15-90-25-BROD/American Association for Cancer Research (American Association for Cancer Research, Inc.)
- U54CA209988/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- PA210068/U.S. Department of Defense (United States Department of Defense)
- U01 CA278923/CA/NCI NIH HHS/United States
- U54 CA209988/CA/NCI NIH HHS/United States
- R01 CA196228/CA/NCI NIH HHS/United States
- U01 CA224012/CA/NCI NIH HHS/United States
- U01CA224012/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U01CA217842/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA212600/CA/NCI NIH HHS/United States
- R21CA263996/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U01 CA217842/CA/NCI NIH HHS/United States
- R01CA212600/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R21 CA263996/CA/NCI NIH HHS/United States
- R01CA196228/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- S10 OD034224/OD/NIH HHS/United States
- U01 CA294548/CA/NCI NIH HHS/United States
- R01 CA186241/CA/NCI NIH HHS/United States
- P30 CA069533/CA/NCI NIH HHS/United States
- R01 CA287672/CA/NCI NIH HHS/United States

