Recent developments and future directions in point-of-care next-generation CRISPR-based rapid diagnosis
- PMID: 39789283
- PMCID: PMC11717804
- DOI: 10.1007/s10238-024-01540-8
Recent developments and future directions in point-of-care next-generation CRISPR-based rapid diagnosis
Abstract
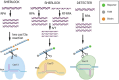
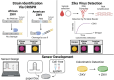
The demand for sensitive, rapid, and affordable diagnostic techniques has surged, particularly following the COVID-19 pandemic, driving the development of CRISPR-based diagnostic tools that utilize Cas effector proteins (such as Cas9, Cas12, and Cas13) as viable alternatives to traditional nucleic acid-based detection methods. These CRISPR systems, often integrated with biosensing and amplification technologies, provide precise, rapid, and portable diagnostics, making on-site testing without the need for extensive infrastructure feasible, especially in underserved or rural areas. In contrast, traditional diagnostic methods, while still essential, are often limited by the need for costly equipment and skilled operators, restricting their accessibility. As a result, developing accessible, user-friendly solutions for at-home, field, and laboratory diagnostics has become a key focus in CRISPR diagnostic innovations. This review examines the current state of CRISPR-based diagnostics and their potential applications across a wide range of diseases, including cancers (e.g., colorectal and breast cancer), genetic disorders (e.g., sickle cell disease), and infectious diseases (e.g., tuberculosis, malaria, Zika virus, and human papillomavirus). Additionally, the integration of machine learning (ML) and artificial intelligence (AI) to enhance the accuracy, scalability, and efficiency of CRISPR diagnostics is discussed, alongside the challenges of incorporating CRISPR technologies into point-of-care settings. The review also explores the potential for these cutting-edge tools to revolutionize disease diagnosis and personalized treatment in the future, while identifying the challenges and future directions necessary to address existing gaps in CRISPR-based diagnostic research.
Keywords: Artificial intelligence; Biosensing technologies; CRISPR-Cas systems; Disease detection; Machine learning; Microfluidic platforms; Nucleic acid diagnostics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests. Ethics approval: Not applicable.
Figures




References
-
- Bhattacharjee G, Gohil N, Lam NL, Singh V. CRISPR-based diagnostics for detection of pathogens. Prog Mol Biol Transl Sci. 2021;181:45–57. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

