Low-avidity T cells drive endogenous tumor immunity in mice and humans
- PMID: 39789375
- PMCID: PMC11785530
- DOI: 10.1038/s41590-024-02044-z
Low-avidity T cells drive endogenous tumor immunity in mice and humans
Abstract
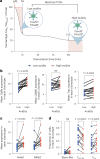
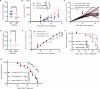
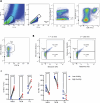
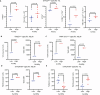
T cells recognize neoepitope peptide-major histocompatibility complex class I on cancer cells. The strength (or avidity) of the T cell receptor-peptide-major histocompatibility complex class I interaction is a critical variable in immune control of cancers. Here, we analyze neoepitope-specific CD8 cells of distinct avidities and show that low-avidity T cells are the sole mediators of cancer control in mice and are solely responsive to checkpoint blockade in mice and humans. High-avidity T cells are ineffective and immune-suppressive. The mechanistic basis of these differences lies in the higher exhaustion status of high-avidity cells. High-avidity T cells have a distinct transcriptomic profile that is used here to calculate an 'avidity score', which we then use for in silico identification of low-avidity and high-avidity T cells in mice and humans. Surprisingly, CD8+ T cells with identical T cell receptors exhibit wide variation in avidities, suggesting an additional level of regulation of T cell activity. Aside from providing a better understanding of endogenous T cell responses to cancer, these findings might instruct future immunotherapy strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.K.S. is a founder of Agenus and a founder, board member and stock owner of Life Science Pharmaceuticals. The other authors declare no competing interests.
Figures









References
-
- Corse, E., Gottschalk, R. A. & Allison, J. P. Strength of TCR–peptide/MHC interactions and in vivo T cell responses. J. Immunol.186, 5039–5045 (2011). - PubMed
-
- Morgan, D. J. et al. Activation of low avidity CTL specific for a self epitope results in tumor rejection but not autoimmunity. J. Immunol.160, 643–651 (1998). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

