OTUD6B regulates KIFC1-dependent centrosome clustering and breast cancer cell survival
- PMID: 39789388
- PMCID: PMC11850729
- DOI: 10.1038/s44319-024-00361-w
OTUD6B regulates KIFC1-dependent centrosome clustering and breast cancer cell survival
Abstract
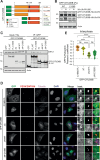
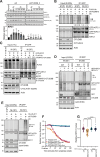
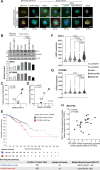
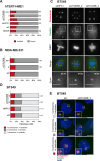
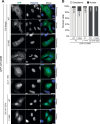
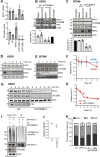
Cancer cells often display centrosome amplification, requiring the kinesin KIFC1/HSET for centrosome clustering to prevent multipolar spindles and cell death. In parallel siRNA screens of deubiquitinase enzymes, we identify OTUD6B as a positive regulator of KIFC1 expression that is required for centrosome clustering in triple-negative breast cancer (TNBC) cells. OTUD6B can localise to centrosomes and the mitotic spindle and interacts with KIFC1. In OTUD6B-deficient cells, we see increased KIFC1 polyubiquitination and premature KIFC1 degradation during mitosis. Depletion of OTUD6B increases multipolar spindles without inducing centrosome amplification. Phenotypic rescue is dependent on OTUD6B catalytic activity and evident upon KIFC1 overexpression. OTUD6B is commonly overexpressed in breast cancer, correlating with KIFC1 protein expression and worse patient survival. TNBC cells with centrosome amplification, but not normal breast epithelial cells, depend on OTUD6B to proliferate. Indeed CRISPR-Cas9 editing results in only OTUD6B-/+ TNBC cells which fail to divide and die. As a deubiquitinase that supports KIFC1 expression, allowing pseudo-bipolar cell division and survival of cancer cells with centrosome amplification, OTUD6B has potential as a novel target for cancer-specific therapies.
Keywords: Centrosome; DUB; Kinesin; Multipolar Spindle; OTU Deubiquitinase 6B.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. Andrew Fielding and Amanda Thomaz declare competing commercial interests due to intellectual property held in field related to this study. The remaining authors declare no competing interests.
Figures






References
-
- Behan FM, Iorio F, Picco G, Goncalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M et al (2019) Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568:511–516 - PubMed
-
- Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V et al (2018) Breast invasive carcinoma TCGA, PanCancer Atlas. Source data from GDC (https://www.cbioportal.org/study?id=brca_tcga_pan_can_atlas_2018) [DATASET]
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

