α-Ketoglutarate dehydrogenase is a therapeutic vulnerability in acute myeloid leukemia
- PMID: 39791576
- PMCID: PMC11969269
- DOI: 10.1182/blood.2024025245
α-Ketoglutarate dehydrogenase is a therapeutic vulnerability in acute myeloid leukemia
Abstract
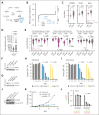
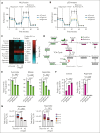
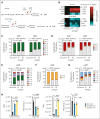
Perturbations in intermediary metabolism contribute to the pathogenesis of acute myeloid leukemia (AML) and can produce therapeutically actionable dependencies. Here, we probed whether α-ketoglutarate (αKG) metabolism represents a specific vulnerability in AML. Using functional genomics, metabolomics, and mouse models, we identified the αKG dehydrogenase complex, which catalyzes the conversion of αKG to succinyl coenzyme A, as a molecular dependency across multiple models of adverse-risk AML. Inhibition of 2-oxoglutarate dehydrogenase (OGDH), the E1 subunit of the αKG dehydrogenase complex, impaired AML progression and drove differentiation. Mechanistically, hindrance of αKG flux through the tricarboxylic acid (TCA) cycle resulted in rapid exhaustion of aspartate pools and blockade of de novo nucleotide biosynthesis, whereas cellular bioenergetics was largely preserved. Additionally, increased αKG levels after OGDH inhibition affected the biosynthesis of other critical amino acids. Thus, this work has identified a previously undescribed, functional link between certain TCA cycle components and nucleotide biosynthesis enzymes across AML. This metabolic node may serve as a cancer-specific vulnerability, amenable to therapeutic targeting in AML and perhaps in other cancers with similar metabolic wiring.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: S.W.L. declares outside consultancy and equity for Oric Pharmaceuticals, Blueprint Medicines, Mirimus, Senecea Therapeutics, Faeth Therapeutics, Selectin Therapeutics, and PMV Pharmaceuticals and outside consultancy (no equity) for Fate Therapeutics (unrelated to this manuscript). Z.S. has received research funds and reagents from Stemline Therapeutics and research reagents from Jazz Pharmaceuticals (neither of which are relevant to this study). The remaining authors declare no competing financial interests.
Figures




Comment in
-
Targeting an AML-sustaining metabolic node.Blood. 2025 Mar 27;145(13):1341-1343. doi: 10.1182/blood.2024027768. Blood. 2025. PMID: 40146158 No abstract available.
References
-
- Dohner H, Wei AH, Appelbaum FR, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345–1377. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

