Methylation of foreign DNA overcomes the restriction barrier of Flavobacterium psychrophilum and allows efficient genetic manipulation
- PMID: 39791877
- PMCID: PMC11837570
- DOI: 10.1128/aem.01448-24
Methylation of foreign DNA overcomes the restriction barrier of Flavobacterium psychrophilum and allows efficient genetic manipulation
Abstract
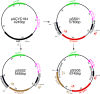
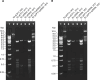
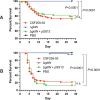
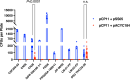
Flavobacterium psychrophilum causes bacterial cold-water disease (BCWD) in salmonids and other fish, resulting in substantial economic losses in aquaculture worldwide. The mechanisms F. psychrophilum uses to cause disease are poorly understood. Despite considerable effort, most strains of F. psychrophilum have resisted attempts at genetic manipulation. F. psychrophilum restriction-modification (R-M) systems may contribute to this resistance. Restriction endonucleases (REases) rapidly degrade nonself DNA if it is not properly methylated by their cognate DNA methyltransferases (MTases). We used comparative genomics to show that R-M systems are abundant in F. psychrophilum and that strain-specific variations partially align with phylogeny. We identified two critical type II R-M systems, HpaII-like (FpsJI) and ScrFI-like (FpsJVI), that are conserved in most of the sequenced strains. Protection of foreign DNA against HpaII and ScrFI was achieved by expression of the MTases M.FpsJI and M.FpsJVI in Escherichia coli. Furthermore, deleting the two REase genes from F. psychrophilum resulted in efficient conjugative DNA transfer from E. coli into the otherwise genetically intractable F. psychrophilum strain CSF259-93. This allowed us to construct a CSF259-93 mutant lacking gldN, a core component of the type IX protein secretion system. The pre-methylation system developed in this study functions in all tested F. psychrophilum strains harboring HpaII-like and ScrFI-like REases. These newly developed genetic tools may allow the identification of key virulence factors and facilitate the development of live attenuated vaccines or other measures to control BCWD.
Importance: Bacterial cold-water disease (BCWD) caused by Flavobacterium psychrophilum is a problem for salmonid aquaculture worldwide, and current control measures are inadequate. An obstacle in understanding and controlling BCWD is that most F. psychrophilum strains resist DNA transfer, thus limiting genetic studies of their virulence mechanisms. F. psychrophilum restriction enzymes that destroy foreign DNA were suspected to contribute to this problem. Here, we used F. psychrophilum DNA methyltransferases to modify and protect foreign DNA from digestion. This allowed efficient conjugative DNA transfer into nine diverse F. psychrophilum strains that had previously resisted DNA transfer. Using this approach, we constructed a gene deletion mutant that failed to cause disease in rainbow trout. Further genetic studies could help determine the molecular factors involved in pathogenesis and may aid development of innovative BCWD control strategies.
Keywords: DNA methyltransferases; DNA transfer; Flavobacterium psychrophilum; bacterial cold-water disease; genetic manipulation; restriction-modification systems; type IX secretion system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- McBride MJ. 2014. The family Flavobacteriaceae, p 643–676. In Rosenberg E, Stackebrandt E, DeLong E, Lory S, Thompson FL (ed), The prokaryotes, 4th ed. Springer.
-
- Calvez S, Navarro-Gonzalez N, Siekoula-Nguedia C, Fournel C, Duchaud E. 2021. High genetic diversity in Flavobacterium psychrophilum isolates from healthy rainbow trout (Oncorhynchus mykiss) farmed in the same watershed, revealed by two typing methods. Appl Environ Microbiol 87:e01398-20. doi: 10.1128/AEM.01398-20 - DOI - PMC - PubMed
-
- Lorenzen E, Dalsgaard IH, Bernardet JF. 1997. Characterization of isolates of Flavobacterium psychrophilum associated with coldwater disease or rainbow trout fry syndrome I: phenotypic and genomic studies. Dis Aquat Org 31:197–208. doi: 10.3354/dao031197 - DOI
-
- Duchaud E, Boussaha M, Loux V, Bernardet J-F, Michel C, Kerouault B, Mondot S, Nicolas P, Bossy R, Caron C, Bessières P, Gibrat J-F, Claverol S, Dumetz F, Le Hénaff M, Benmansour A. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat Biotechnol 25:763–769. doi: 10.1038/nbt1313 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

