Modulation of cytokeratin and cytokine/chemokine expression following influenza virus infection of differentiated human tonsillar epithelial cells
- PMID: 39791909
- PMCID: PMC11852761
- DOI: 10.1128/jvi.01460-24
Modulation of cytokeratin and cytokine/chemokine expression following influenza virus infection of differentiated human tonsillar epithelial cells
Abstract
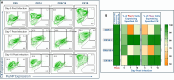
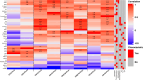
The tonsils have been identified as a site of replication for Epstein-Barr virus, adenovirus, human papillomavirus, and other respiratory viruses. Human tonsil epithelial cells (HTECs) are a heterogeneous group of actively differentiating cells. Here, we investigated the cellular features and susceptibility of differentiated HTECs to specific influenza viruses, including expression of avian-type and mammalian-type sialic acid (SA) receptors, viral replication dynamics, and the associated cytokine secretion profiles. We found that differentiated HTECs possess more abundant α2,3-linked SA (preferentially bound by avian influenza viruses) than α2,6-linked SA (preferentially bound by mammalian strains). This dual receptor expression suggests a role in influenza virus adaptation and tropism within the tonsils by facilitating the binding and entry of multiple influenza virus strains. Our results indicated the susceptibility of differentiated HTECs to a wide range of influenza viruses from human, swine, and avian hosts. Virus production for most strains was detected as early as 1 day post-infection (dpi), and typically peaked by 3 dpi. However, pandemic H1N1 virus showed remarkably delayed replication kinetics that did not peak until at least 7 dpi. Notably, influenza virus infection impacted the expression of cytokeratins in HTEC cultures, which correlated with altered cytokine secretion patterns. These patterns varied within the strains but were most distinct in swine H3N2 infection. In conclusion, differentiated HTECs exhibited a strain-specific pattern of influenza virus replication and innate immune responses that included changes in cytokeratin and cytokine expression. These studies shed light on the complex interplay between influenza viruses and host cells in the tonsils.
Importance: To develop effective interventions against influenza, it is important to identify host factors affecting pathogenesis and immune responses. Tonsils are lymphoepithelial organs characterized by infiltration of B and T lymphocytes into the squamous epithelium of tonsillar crypts, beneath which germinal centers play key roles in antigen processing and the immune response. Influenza virus tropism in the human upper respiratory tract is a key determinant of host-range, pathogenesis, and transmission. Accordingly, experimental models using primary cells from the human respiratory tract are relevant for assessing virus tropism and replication competence. Our study addresses the dynamics of influenza virus replication in HTECs, including cellular tropism, infectivity, and cytokeratin and cytokine expression. The results of this study highlight the complex interplay between structural proteins and immune signaling pathways, all of which provide valuable insights into host-virus interactions.
Keywords: chemokines; crypts; cytokeratins, keratin; cytokines; influenza viruses; reticular epithelial cells; sialic acid receptors; squamous epithelial cells; tonsils; tropism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- CDC . 2019. Disease burden of influenza. Available from: https://www.cdc.gov/flu/about/burden/index.html https://www.cdc.gov/flu/...
-
- World Health Organization . 2016. Tool for influenza pandemic risk assessment. Influenza Pandemic Risk Assessment.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

