Exceptional Uptake, Limited Protein Expression: Liver Macrophages Lost in Translation of Synthetic mRNA
- PMID: 39792811
- PMCID: PMC11884593
- DOI: 10.1002/advs.202409729
Exceptional Uptake, Limited Protein Expression: Liver Macrophages Lost in Translation of Synthetic mRNA
Abstract
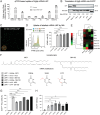
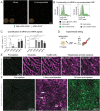
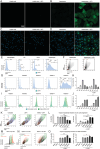
Most gene therapies exert their actions via manipulation of hepatocytes (parenchymal cells) and the reasons behind the suboptimal performance of synthetic mRNA in non-parenchymal cells (NPC) such as Kupffer cells (KC), and liver macrophages, remain unclear. Here, the spatio-temporal distribution of mRNA encoding enhanced green fluorescent protein (Egfp), siRNA, or both co-encapsulated into lipid nanoparticles (LNP) in the liver in vivo using real-time intravital imaging is investigated. Although both KC and hepatocytes demonstrate comparable high and rapid uptake of mRNA-LNP and siRNA-LNP in vivo, the translation of Egfp mRNA occurs exclusively in hepatocytes during intravital imaging. Despite attempts such as inhibiting intracellular ribonuclease, substituting uridine bases in mRNA with pseudouridine, and using a different ionizable lipid in the LNP mixture, no substantial increase in Egfp translation by NPC is possible. The investigation reveals that hepatocytes, which are distinct from other liver cells due to their polyploidy, exhibit significantly elevated levels of total RNA and protein, along with a higher proportion of ribosomal protein per individual cell. Consequently, fundamental cellular differences account for the low mRNA translation observed in NPC. The findings therefore suggest that cellular biology imposes a natural limitation on synthetic mRNA translation that is strongly influenced by cellular ploidy.
Keywords: hepatocytes; lipid nanoparticles; non‐parenchymal cells; ploidy; synthetic mRNA.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- a) Woitok M. M., Zoubek M. E., Doleschel D., Bartneck M., Mohamed M. R., Kiessling F., Lederle W., Trautwein C., Cubero F. J., Cell Death Dis. 2020, 11, 343; - PMC - PubMed
- b) Lin C., Jans A., Wolters J. C., Mohamed M. R., Van der Vorst E. P. C., Trautwein C., Bartneck M., Adv. Healthcare Mater. 2024, 13, e2202670; - PubMed
- c) Mohamed M. R., Haybaeck J., Wu H., Su H., Bartneck M., Lin C., Boekschoten M. V., Boor P., Goeppert B., Rupp C., Strnad P., Davis R. J., Cubero F. J., Trautwein C., JHEP Rep. 2023, 5, 100854. - PMC - PubMed
-
- Kulkarni J. A., Witzigmann D., Thomson S. B., Chen S., Leavitt B. R., Cullis P. R., van der Meel R., Nat. Nanotechnol. 2021, 16, 630. - PubMed
-
- Heymann F., Peusquens J., Ludwig‐Portugall I., Kohlhepp M., Ergen C., Niemietz P., Martin C., van Rooijen N., Ochando J. C., Randolph G. J., Luedde T., Ginhoux F., Kurts C., Trautwein C., Tacke F., Hepatology 2015, 62, 279. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
