High-throughput drug screening identifies SMAC mimetics as enhancers of NK-cell cytotoxicity in chronic myeloid leukemia
- PMID: 39792962
- PMCID: PMC12000656
- DOI: 10.1182/blood.2024025286
High-throughput drug screening identifies SMAC mimetics as enhancers of NK-cell cytotoxicity in chronic myeloid leukemia
Abstract
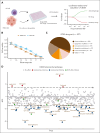
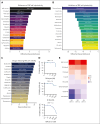
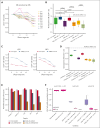
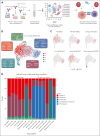
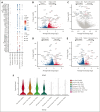
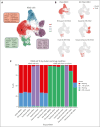
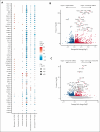
Natural killer (NK) cells have proven to be safe and effective immunotherapies, associated with favorable treatment responses in chronic myeloid leukemia (CML). Augmenting NK-cell function with oncological drugs could improve NK-cell-based immunotherapies. Here, we used a high-throughput drug screen consisting of >500 small-molecule compounds, to systematically evaluate the effects of oncological drugs on primary NK cells against CML cells. We identified second mitochondrially derived activator of caspases (SMAC) mimetics as potent enhancers of NK-cell cytotoxicity in both cell lines and primary patient samples. In contrast, several drug classes, including glucocorticoids and tyrosine kinase inhibitors such as dasatinib, inhibited NK-cell cytotoxicity. Single-cell RNA sequencing revealed drug-induced transcriptomic changes in both NK and target CML cells. SMAC mimetics upregulated NF-κB target genes in NK cells, potentially contributing to their enhanced cytotoxicity. Inhibitory drugs dexamethasone, dasatinib, and sotrastaurin prevented NK-cell transition to an activated state and suppressed the expression of interferon gamma (IFN-γ) by NK cells, thus preventing IFN-γ-mediated target cell transcriptomic response. In conclusion, we discovered that SMAC mimetics sensitize cancer cells to NK-cell-mediated killing, with potential clinical applications especially in patients with advanced phase CML.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: S.M. has received honoraria and research funding from Novartis, Pfizer, and Bristol Myers Squibb; and honoraria from Dren Bio (not related to this study). H.H.-H., on behalf of the Nordic CML study group, has received research funding from Bristol Myers Squibb, Novartis, Incyte, Pfizer, and Austrian Orphan Pharma, unrelated to this study; and has received lecture fees from Incyte. O.D. has received research funding from Gilead Sciences and Incyte, unrelated to this work; and personal fees from Sanofi, unrelated to this work. The remaining authors declare no competing financial interests.
A complete list of the members of the iCAN Study Group appears in the supplemental Appendix.
Figures



Comment in
-
SMAC mimetics in action in chronic myeloid leukemia.Blood. 2025 Apr 10;145(15):1596-1597. doi: 10.1182/blood.2024027994. Blood. 2025. PMID: 40208653 No abstract available.
References
-
- Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph' translocation. Nature. 1985;315(6022):758–761. - PubMed
-
- Mahon F-X, Réa D, Guilhot J, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029–1035. - PubMed
-
- Graham SM, Jørgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–325. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

