Gonadal sex and temperature independently influence germ cell differentiation and meiotic progression in Trachemys scripta
- PMID: 39793067
- PMCID: PMC11725912
- DOI: 10.1073/pnas.2413191121
Gonadal sex and temperature independently influence germ cell differentiation and meiotic progression in Trachemys scripta
Abstract
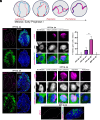
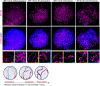
In species with genetic sex determination (GSD), the sex identity of the soma determines germ cell fate. For example, in mice, XY germ cells that enter an ovary differentiate as oogonia, whereas XX germ cells that enter a testis initiate differentiation as spermatogonia. However, numerous species lack a GSD system and instead display temperature-dependent sex determination (TSD). In the red-eared slider turtle, Trachemys scripta, a TSD model species with a warm female promoting temperature (FPT) and cool male promoting temperature (MPT) system, temperature directly affects germ cell number. In this study, we examined whether temperature directly affects other aspects of germ cell differentiation/sex identity. We uncoupled temperature and the sexual fate of the gonad by incubating eggs at MPT and treating with 17β-estradiol, a scheme that invariably produces ovaries. Through analysis of meiotic spreads, we showed that germ cells in FPT ovaries follow the typical pattern of initiating meiosis and progress through prophase I. However, in E2-induced ovaries that incubated at MPT, germ cells entered prophase I yet fail to exhibit synapsis. These results, combined with our single-cell transcriptome analysis, reveal a direct effect of temperature on germ cell sexual differentiation independent of its effect on the gonadal soma. These results imply that not all events of meiosis are under somatic control, at least not in this TSD species.
Keywords: germ cell; meiosis; sex determination.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



Similar articles
-
Estrogen receptor and temperature independently influence sex determination in the red-eared slider turtle.Front Endocrinol (Lausanne). 2025 Jul 21;16:1632672. doi: 10.3389/fendo.2025.1632672. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40761814 Free PMC article.
-
Histological and immunohistochemical analysis of gonadal development in the veiled chameleon (Chamaeleo calyptratus).Anat Rec (Hoboken). 2025 Sep;308(9):2490-2507. doi: 10.1002/ar.25621. Epub 2024 Dec 24. Anat Rec (Hoboken). 2025. PMID: 39719868
-
Transcriptome analysis reveals the sex-switching mechanism of juvenile hermaphroditism in silver pomfret (Pampus argenteus).Biol Sex Differ. 2025 Jul 14;16(1):51. doi: 10.1186/s13293-025-00736-1. Biol Sex Differ. 2025. PMID: 40660379 Free PMC article.
-
Psychological interventions for adults who have sexually offended or are at risk of offending.Cochrane Database Syst Rev. 2012 Dec 12;12(12):CD007507. doi: 10.1002/14651858.CD007507.pub2. Cochrane Database Syst Rev. 2012. PMID: 23235646 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
Cited by
-
Estrogen receptor and temperature independently influence sex determination in the red-eared slider turtle.Front Endocrinol (Lausanne). 2025 Jul 21;16:1632672. doi: 10.3389/fendo.2025.1632672. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40761814 Free PMC article.
-
Clonal analysis reveals differential PGC contributions to the early germline and ovarian reserve.bioRxiv [Preprint]. 2025 May 7:2025.04.30.651497. doi: 10.1101/2025.04.30.651497. bioRxiv. 2025. PMID: 40654649 Free PMC article. Preprint.
References
-
- Berta P., et al. , Genetic evidence equating SRY and the testis-determining factor. Nature 348, 448–450 (1990). - PubMed
-
- Koopman P., Münsterberg A., Capel B., Vivian N., Lovell-Badge R., Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348, 450–452 (1990). - PubMed
-
- Capel B., Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 18, 675–689 (2017). - PubMed
-
- McLaren A., Germ cells and germ cell sex. Philos Trans. R Soc. Lond. B Biol. Sci. 350, 229–233 (1995). - PubMed
MeSH terms
Grants and funding
- F32HD113220/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- F32 HD113220/HD/NICHD NIH HHS/United States
- EMBO ALTF 537-2020/European Molecular Biology Organization (EMBO)
- R01HD103064/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 HD103064/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources

