Genesis and regulation of C-terminal cyclic imides from protein damage
- PMID: 39793072
- PMCID: PMC11725857
- DOI: 10.1073/pnas.2415976121
Genesis and regulation of C-terminal cyclic imides from protein damage
Abstract
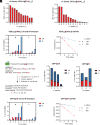
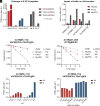
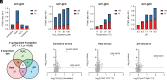
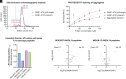
C-Terminal cyclic imides are posttranslational modifications that can arise from spontaneous intramolecular cleavage of asparagine or glutamine residues resulting in a form of irreversible protein damage. These protein damage events are recognized and removed by the E3 ligase substrate adapter cereblon (CRBN), indicating that these aging-related modifications may require cellular quality control mechanisms to prevent deleterious effects. However, the factors that determine protein or peptide susceptibility to C-terminal cyclic imide formation or their effect on protein stability have not been explored in detail. Here, we characterize the primary and secondary structures of peptides and proteins that promote intrinsic formation of C-terminal cyclic imides in comparison to deamidation, a related form of protein damage. Extrinsic effects from solution properties and stressors on the cellular proteome additionally promote C-terminal cyclic imide formation on proteins like glutathione synthetase that are susceptible to aggregation if the protein damage products are not removed by CRBN. This systematic investigation provides insight into the regions of the proteome that are prone to these unexpectedly frequent modifications, the effects of this form of protein damage on protein stability, and the biological role of CRBN.
Keywords: cereblon; deamidation; post-translational modification; protein aggregation; protein damage.
Conflict of interest statement
Competing interests statement:Harvard University has filed a PCT patent application on April 13, 2022 covering the chemical structures and their use. C.M.W. and W.X. are inventors of this patent. The Woo lab receives or has received support from Merck and Ono Pharmaceuticals.
Figures



Update of
-
Genesis and regulation of C-terminal cyclic imides from protein damage.bioRxiv [Preprint]. 2024 Aug 10:2024.08.09.606997. doi: 10.1101/2024.08.09.606997. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2415976121. doi: 10.1073/pnas.2415976121. PMID: 39211211 Free PMC article. Updated. Preprint.
References
-
- Harmel R., Fiedler D., Features and regulation of non-enzymatic post-translational modifications. Nat. Chem. Biol. 14, 244–252 (2018). - PubMed
-
- Brownlee M., Vlassara H., Cerami A., Nonenzymatic glycosylation and the pathogenesis of diabetic complications. Ann. Intern. Med. 101, 527–537 (1984). - PubMed
-
- Perler F. B., Xu M. Q., Paulus H., Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1, 292–299 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

