A conifer metabolite corrects episodic ataxia type 1 by voltage sensor-mediated ligand activation of Kv1.1
- PMID: 39793113
- PMCID: PMC11745346
- DOI: 10.1073/pnas.2411816122
A conifer metabolite corrects episodic ataxia type 1 by voltage sensor-mediated ligand activation of Kv1.1
Abstract
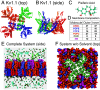
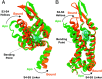
Loss-of-function sequence variants in KCNA1, which encodes the voltage-gated potassium channel Kv1.1, cause Episodic Ataxia Type 1 (EA1) and epilepsy. Due to a paucity of drugs that directly rescue mutant Kv1.1 channel function, current therapeutic strategies for KCNA1-linked disorders involve indirect modulation of neuronal excitability. Native Americans have traditionally used conifer extracts to treat paralysis, weakness, and pain, all of which may involve altered electrical activity and/or Kv1.1 dysfunction specifically. Here, screening conifer extracts, we found that Chamaecyparis pisifera increases wild-type (WT) Kv1.1 activity, as does its prominent metabolite, the abietane diterpenoid pisiferic acid. Uniquely, pisiferic acid also restored function in 12/12 EA1-linked mutant Kv1.1 channels tested in vitro. Crucially, pisiferic acid (1 mg/kg) restored WT function in Kv1.1E283K/+ mice, a model of human EA1. Experimentally validated all-atom molecular dynamics simulations in a neuron-like membrane revealed that the Kv1.1 voltage-sensing domain (VSD) also acts as a ligand-binding domain akin to those of classic ligand-gated channels; binding of pisiferic acid induces a conformational shift in the VSD that ligand-dependently opens the pore. Conifer metabolite pisiferic acid is a promising and versatile therapeutic lead for EA1 and other Kv1.1-linked disorders.
Keywords: KCNA1; Kv1.1; episodic ataxia; potassium channel; voltage gating.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures








References
-
- Hasan S. M., D’Adamo M. C., “Episodic Ataxia Type 1” in GeneReviews((R)), Adam M. P., et al., Eds. (University of Washington, Seattle, WA, 1993).
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

