Small-molecule-induced ERBB4 activation to treat heart failure
- PMID: 39794341
- PMCID: PMC11724075
- DOI: 10.1038/s41467-024-54908-5
Small-molecule-induced ERBB4 activation to treat heart failure
Abstract
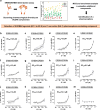
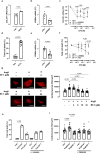
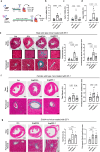
Heart failure is a common and deadly disease requiring new treatments. The neuregulin-1/ERBB4 pathway offers cardioprotective benefits, but using recombinant neuregulin-1 as therapy has limitations due to the need for intravenous delivery and lack of receptor specificity. We hypothesize that small-molecule activation of ERBB4 could protect against heart damage and fibrosis. To test this, we conduct a screening of 10,240 compounds and identify eight structurally similar ones (EF-1 to EF-8) that induce ERBB4 dimerization, with EF-1 being the most effective. EF-1 reduces cell death and hypertrophy in cardiomyocytes and decreases collagen production in cardiac fibroblasts in an ERBB4-dependent manner. In wild-type mice, EF-1 inhibits angiotensin-II-induced fibrosis in males and females and reduces heart damage caused by doxorubicin and myocardial infarction in females, but not in Erbb4-null mice. This study shows that small-molecule ERBB4 activation is feasible and may lead to a novel class of drugs for treating heart failure.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Patent “MODULATORS OF ERBB4 IN THE TREATMENT OF DISEASES”; EP20210160742; Inventors: V.F.M.S., G.W.D.K., E.F., H.D.W.
Figures




References
-
- Gassmann, M. et al. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature378, 390–394 (1995). - PubMed
-
- Meyer, D. & Birchmeier, C. Multiple essential functions of neuregulin in development. Nature378, 386–390 (1995). - PubMed
-
- Lee, K. F. et al. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature378, 394–398 (1995). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

