Compare Analysis of Codon Usage Bias of Nuclear Genome in Eight Sapindaceae Species
- PMID: 39795897
- PMCID: PMC11720230
- DOI: 10.3390/ijms26010039
Compare Analysis of Codon Usage Bias of Nuclear Genome in Eight Sapindaceae Species
Abstract
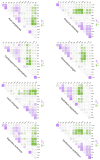
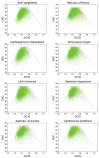
Codon usage bias (CUB) refers to the different frequencies with which various codons are utilized within a genome. Examining CUB is essential for understanding genome structure, function, and evolution. However, little was known about codon usage patterns and the factors influencing the nuclear genomes of eight ecologically significant Sapindaceae species widely utilized for food and medicine. In this study, an analysis of nucleotide composition revealed a higher A/T content and showed a preference for A/T at the third codon position in the eight species of Sapindaceae. A correspondence analysis of relative synonymous codon usage explained only part of the variation, suggesting that not only natural selection but also various other factors contribute to selective constraints on codon bias in the nuclear genomes of the eight Sapindaceae species. Additionally, ENC-GC3 plot, PR2-Bias, and neutrality plot analyses indicated that natural selection exerted a greater influence than mutation pressure across these eight species. Among the eight Sapindaceae species, 16 to 26 optimal codons were identified, with two common high-frequency codons: AGA (encoding Arg) and GCU (encoding Ala). The clustering heat map, which included the 8 Sapindaceae species and 13 other species, revealed two distinct clusters corresponding to monocots and dicots. This finding suggested that CUB analysis was particularly effective in elucidating evolutionary relationships at the family level. Collectively, our results emphasized the distinct codon usage characteristics and unique evolutionary traits of the eight Sapindaceae species.
Keywords: Sapindaceae; codon usage bias; evolution; mutation; nature selection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

