Identification of Kunitz-Type Inhibitor Gene Family of Populus yunnanensis Reveals a Stress Tolerance Function in Inverted Cuttings
- PMID: 39796046
- PMCID: PMC11720115
- DOI: 10.3390/ijms26010188
Identification of Kunitz-Type Inhibitor Gene Family of Populus yunnanensis Reveals a Stress Tolerance Function in Inverted Cuttings
Abstract
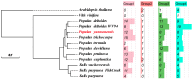
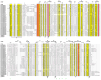
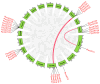
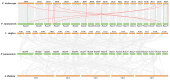
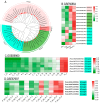
Plant protease inhibitors are a ubiquitous feature of plant species and exert a substantial influence on plant stress responses. However, the KTI (Kunitz trypsin inhibitor) family responding to abiotic stress has not been fully characterized in Populus yunnanensis. In this study, we conducted a genome-wide study of the KTI family and analyzed their gene structure, gene duplication, conserved motifs, cis-acting elements, and response to stress treatment. A total of 29 KTIs were identified in the P. yunnanensis genome. Based on phylogenetic analysis, the PyKTIs were divided into four groups (1,2, 3, and 4). Promoter sequence analysis showed that the PyKTIs contain many cis-acting elements related to light, plant growth, hormone, and stress responses, indicating that PyKTIs are widely involved in various biological regulatory processes. RNA sequencing and real-time quantitative polymerase chain reaction analysis showed that KTI genes were differentially expressed under the inverted cutting stress of P. yunnanensis. Transcriptome analysis of P. yunnanensis leaves revealed that PyKTI16, PyKTI18, and PyKTI19 were highly upregulated after inverted cutting. Through the GEO query of Populus transcriptome data, KTI genes played a positive defense role in MeJa, drought, time series, and pathogen stress. This study provided comprehensive information for the KTI family in P. yunnanensis, which should be helpful for the functional characterization of P. yunnanensis KTI genes in the future.
Keywords: KTI gene family; Populus yunnanensis; inverted cuttings; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

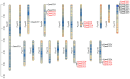





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

