Head-to-Head Comparison of Aptamer- and Antibody-Based Proteomic Platforms in Human Cerebrospinal Fluid Samples from a Real-World Memory Clinic Cohort
- PMID: 39796148
- PMCID: PMC11720409
- DOI: 10.3390/ijms26010286
Head-to-Head Comparison of Aptamer- and Antibody-Based Proteomic Platforms in Human Cerebrospinal Fluid Samples from a Real-World Memory Clinic Cohort
Abstract
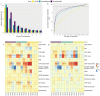
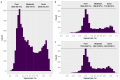
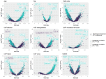
High-throughput proteomic platforms are crucial to identify novel Alzheimer's disease (AD) biomarkers and pathways. In this study, we evaluated the reproducibility and reliability of aptamer-based (SomaScan® 7k) and antibody-based (Olink® Explore 3k) proteomic platforms in cerebrospinal fluid (CSF) samples from the Ace Alzheimer Center Barcelona real-world cohort. Intra- and inter-platform reproducibility were evaluated through correlations between two independent SomaScan® assays analyzing the same samples, and between SomaScan® and Olink® results. Association analyses were performed between proteomic measures, CSF biological traits, sample demographics, and AD endophenotypes. Our 12-category metric of reproducibility combining correlation analyses identified 2428 highly reproducible SomaScan CSF measures, with over 600 proteins well reproduced on another proteomic platform. The association analyses among AD clinical phenotypes revealed that the significant associations mainly involved reproducible proteins. The validation of reproducibility in these novel proteomics platforms, measured using this scarce biomaterial, is essential for accurate analysis and proper interpretation of innovative results. This classification metric could enhance confidence in multiplexed proteomic platforms and improve the design of future panels.
Keywords: Alzheimer’s disease; Olink; SomaScan; biomarkers; cerebrospinal fluid; mild cognitive impairment; proteomics.
Conflict of interest statement
Authors Asif Khan, Bart Smets and Alfredo Cabrera-Socorro are employed by the Janssen Pharmaceutica NV, a Johnson & Johnson Company. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S.B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G., et al. Homogenous 96-Plex PEA Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability. PLoS ONE. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CNV-304-PRF-866/CIBERNED employment plan
- 01EK2102B/German Research Foundation (DFG)
- AC19/00097 and 01ED2007A/Joint Program for Neurodegenerative Diseases (JPND) grant
- R01 AG074007/AG/NIA NIH HHS/United States
- FI20/00215/Instituto de Salud Carlos III
- PR067/21/Agency for Innovation and Entrepreneurship (VLAIO) grant
- PID2021-122473OA-I00, PID2021-123462OB-I00 and PID2019-106625RB-I00/Spanish Ministry of Science and Innovation, Proyectos de Generación de Conocimiento grants
- CB06/05/2004 and CB18/05/00010/Biomedical Research Networking Center on Neurodegenerative Diseases
- 115975/EFPIA Innovative Medicines Initiative Joint Undertaking Grant
- CD22/00125/Sara Borrell grant (ISCIII)
- PI17/01474, PI19/00335, PI22/01403 and PI22/00258/Instituto de Salud Carlos III
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

