Evaluation of the Anti-Amyloid and Anti-Inflammatory Properties of a Novel Vanadium(IV)-Curcumin Complex in Lipopolysaccharides-Stimulated Primary Rat Neuron-Microglia Mixed Cultures
- PMID: 39796150
- PMCID: PMC11720140
- DOI: 10.3390/ijms26010282
Evaluation of the Anti-Amyloid and Anti-Inflammatory Properties of a Novel Vanadium(IV)-Curcumin Complex in Lipopolysaccharides-Stimulated Primary Rat Neuron-Microglia Mixed Cultures
Abstract
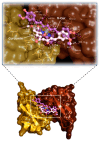
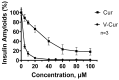
Lipopolysaccharides (LPS) are bacterial mediators of neuroinflammation that have been detected in close association with pathological protein aggregations of Alzheimer's disease. LPS induce the release of cytokines by microglia and mediate the upregulation of inducible nitric oxide synthase (iNOS)-a mechanism also associated with amyloidosis. Curcumin is a recognized natural medicine but has extremely low bioavailability. V-Cur, a novel hemocompatible Vanadium(IV)-curcumin complex with higher solubility and bioactivity than curcumin, is studied here. Co-cultures consisting of rat primary neurons and microglia were treated with LPS and/or curcumin or V-Cur. V-Cur disrupted LPS-induced overexpression of amyloid precursor protein (APP) and the in vitro aggregation of human insulin (HI), more effectively than curcumin. Cell stimulation with LPS also increased full-length, inactive, and total iNOS levels, and the inflammation markers IL-1β and TNF-α. Both curcumin and V-Cur alleviated these effects, with V-Cur reducing iNOS levels more than curcumin. Complementary insights into possible bioactivity mechanisms of both curcumin and V-Cur were provided by In silico molecular docking calculations on Aβ1-42, APP, Aβ fibrils, HI, and iNOS. This study renders curcumin-based compounds a promising anti-inflammatory intervention that may be proven a strong tool in the effort to mitigate neurodegenerative disease pathology and neuroinflammatory conditions.
Keywords: amyloid precursor protein; curcumin; lipopolysaccharides; mixed neuron-glia cultures; neuroinflammation; vanadium-curcumin complex.
Conflict of interest statement
The authors have no competing interests to declare that are relevant to the content of this article.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

