Molecular Basis of Oncogenic PI3K Proteins
- PMID: 39796708
- PMCID: PMC11720314
- DOI: 10.3390/cancers17010077
Molecular Basis of Oncogenic PI3K Proteins
Abstract
The dysregulation of phosphatidylinositol 3-kinase (PI3K) signaling plays a pivotal role in driving neoplastic transformation by promoting uncontrolled cell survival and proliferation. This oncogenic activity is primarily caused by mutations that are frequently found in PI3K genes and constitutively activate the PI3K signaling pathway. However, tumorigenesis can also arise from nonmutated PI3K proteins adopting unique active conformations, further complicating the understanding of PI3K-driven cancers. Recent structural studies have illuminated the functional divergence among highly homologous PI3K proteins, revealing how subtle structural alterations significantly impact their activity and contribute to tumorigenesis. In this review, we summarize current knowledge of Class I PI3K proteins and aim to unravel the complex mechanism underlying their oncogenic traits. These insights will not only enhance our understanding of PI3K-mediated oncogenesis but also pave the way for the design of novel PI3K-based therapies to combat cancers driven by this signaling pathway.
Keywords: PI3K; oncogenic mutation; oncogenic transformation; protein structure.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures
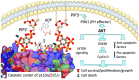








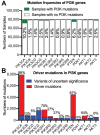
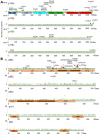
References
-
- Pacold M.E., Suire S., Perisic O., Lara-Gonzalez S., Davis C.T., Walker E.H., Hawkins P.T., Stephens L., Eccleston J.F., Williams R.L. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/S0092-8674(00)00196-3. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

